Bacterial proteases participate in the proteolytic elimination of misfolded or aggregated proteins, carried out by members of the AAA+ protein superfamily such as Hsp100/Clp (heat shock protein-100/caseinolytic protease), Lon, and FtsH. It is estimated that the Clp and Lon families perform around 80% of cellular proteolysis in bacteria. The HSP100/Clp family of ATPases plays crucial roles in the folding, assembly, and degradation of proteins during normal growth and, mainly, under stress-inducing conditions. This family is formed by several ATPase chaperones and the peptidase ClpP (caseinolytic protease proteolytic subunit).
- Clp proteases
- Gram-positive
- chaperone-protease complex
1. Introduction
2. Clp Protease Families
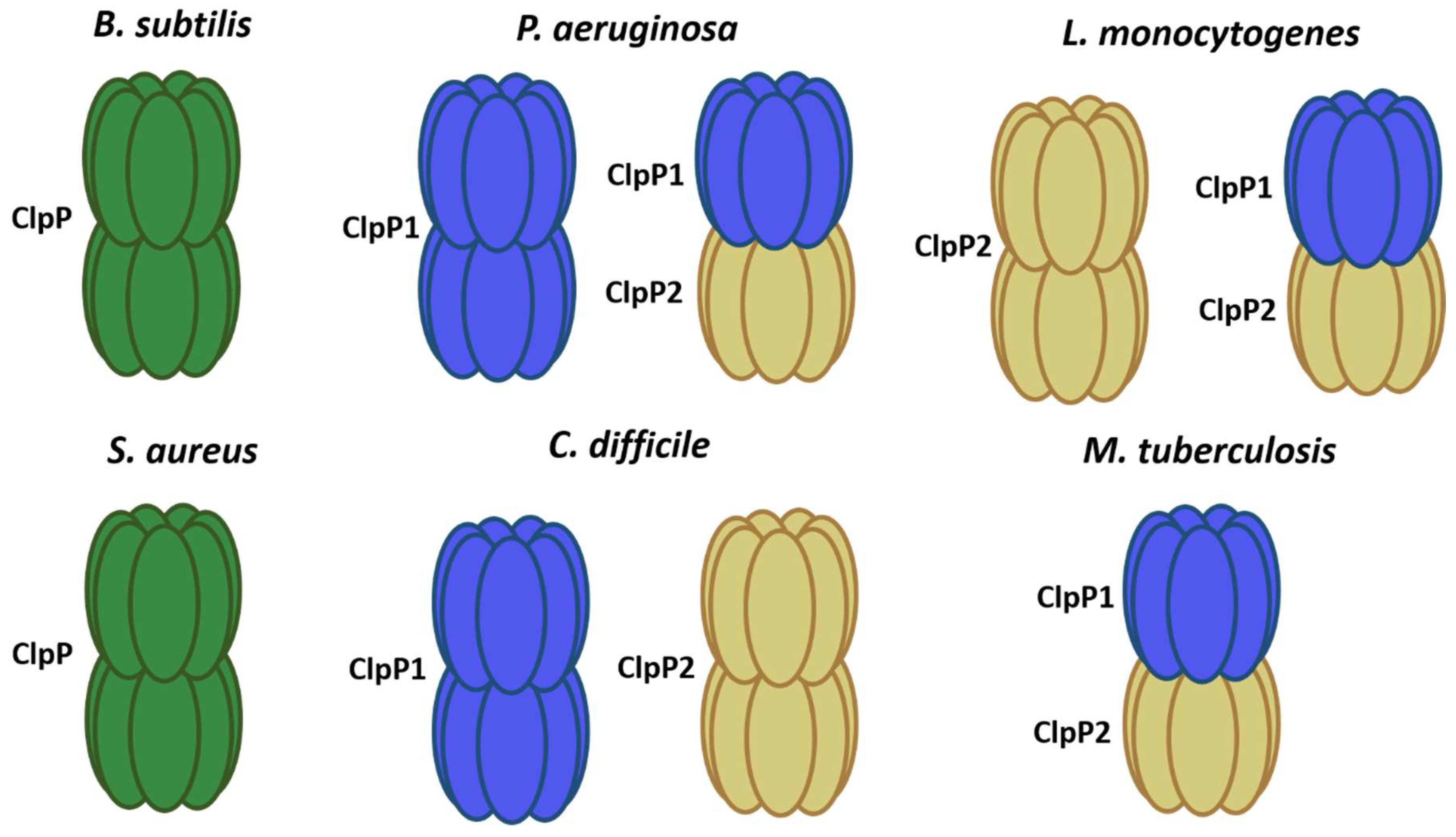
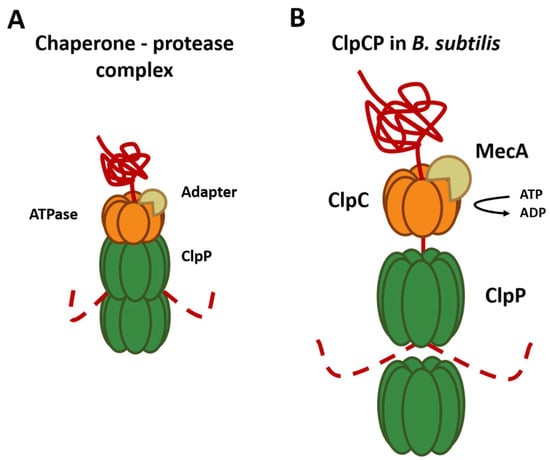
Regulation of Complex
This entry is adapted from the peer-reviewed paper 10.3390/bacteria2010002
References
- Bhandari, V.; Wong, K.S.; Zhou, J.L.; Mabanglo, M.F.; Batey, R.A.; Houry, W.A. The Role of ClpP Protease in Bacterial Pathogenesis and Human Diseases. ACS Chem. Biol. 2018, 13, 1413–1425.
- Striebel, F.; Kress, W.; Weber-Ban, E. Controlled destruction: AAA ATPases in protein degradation from bacteria to eukaryotes. Curr. Opin. Struct. Biol. 2009, 19, 209–217.
- Maurizi, M.R.; Thompson, M.W.; Singh, S.K.; Kim, S. Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods Enzymol. Proteolytic Enzym. Serine Cysteine Pept. 1994, 244, 314–331.
- Goldberg, A.L.; Moerschell, R.P.; Hachung, C.; Maurizi, M.R. ATP-dependent protease La (Lon) from Escherichia coli. Methods Enzymol. Proteolytic Enzym. Serine Cysteine Pept. 1994, 244, 350–375.
- Frees, D.; Savijoki, K.; Varmanen, P.; Ingmer, H. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 2007, 63, 1285–1295.
- Kirstein, J.; Molière, N.; Dougan, D.A.; Turgay, K. Adapting the machine: Adaptor proteins for Hsp100/Clp and AAA proteases. Nat. Rev. Microbiol. 2009, 7, 589–599.
- Brötz-Oesterhelt, H.; Beyer, D.; Kroll, H.P.; Endermann, R.; Ladel, C.; Schroeder, W.; Hinzen, B.; Raddatz, S.; Paulsen, H.; Henninger, K.; et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 2005, 11, 1082–1087.
- Molière, N.; Hoßmann, J.; Schäfer, H.; Turgay, K. Role of Hsp100/Clp Protease Complexes in Controlling the Regulation of Motility in Bacillus subtilis. Front. Microbiol. 2016, 7, 315.
- Kress, W.; Mutschler, H.; Weber-Ban, E. Both ATPase Domains of ClpA Are Critical for Processing of Stable Protein Structures. J. Biol. Chem. 2009, 284, 31441–31452.
- Yu, Y.; Yan, F.; He, Y.; Qin, Y.; Chen, Y.; Chai, Y.; Guo, J.H. The ClpY-ClpQ protease regulates multicellular development in Bacillus subtilis. Microbiology 2018, 164, 848–862.
- Schirmer, E.C.; Glover, J.R.; Singer, M.A.; Lindquist, S. HSP100/Clp proteins: A common mechanism explains diverse functions. Trends Biochem. Sci. 1996, 21, 289–296.
- Lemos, J.A.; Burne, R.A. Regulation and Physiological Significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 2002, 184, 6357–6366.
- Gottesman, S.; Maurizi, M.R.; Wickner, S. Regulatory Subunits of Energy-Dependent Proteases. Cell 1997, 91, 435–438.
- Kruger, E. Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 2001, 20, 852–863.
- Moreno-Cinos, C.; Goossens, K.; Salado, I.G.; Veken, P.V.; Winter, H.D.; Augustyns, K. ClpP Protease, a Promising Antimicrobial Target. Int. J. Mol. Sci. 2019, 20, 2232.
- Frees, D.; Thomsen, L.E.; Ingmer, H. Staphylococcus aureus ClpYQ plays a minor role in stress survival. Arch. Microbiol. 2005, 183, 286–291.
- Kruger, E.; Witt, E.; Ohlmeier, S.; Hanschke, R.; Hecker, M. The Clp Proteases of Bacillus subtilis Are Directly Involved in Degradation of Misfolded Proteins. J. Bacteriol. 2000, 182, 3259–3265.
- Akopian, T.; Kandror, O.; Raju, R.M.; Unnikrishnan, M.; Rubin, E.J.; Goldberg, A.L. The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J. 2012, 31, 1529–1541.
- Hall, B.M.; Breidenstein, E.B.; Fuente-Núñez, C.D.; Reffuveille, F.; Mawla, G.D.; Hancock, R.E.; Baker, T.A. Two Isoforms of Clp Peptidase in Pseudomonas aeruginosa Control Distinct Aspects of Cellular Physiology. J. Bacteriol. 2017, 199, e00568-16.
- Lavey, N.P.; Shadid, T.; Ballard, J.D.; Duerfeldt, A.S. Clostridium difficile ClpP Homologues are Capable of Uncoupled Activity and Exhibit Different Levels of Susceptibility to Acyldepsipeptide Modulation. ACS Infect. Dis. 2019, 5, 79–89.
- Olivares, A.O.; Baker, T.A.; Sauer, R.T. Mechanical Protein Unfolding and Degradation. Annu. Rev. Physiol. 2018, 80, 413–429.
- Gerth, U.; Kock, H.; Kusters, I.; Michalik, S.; Switzer, R.L.; Hecker, M. Clp-Dependent Proteolysis Down-Regulates Central Metabolic Pathways in Glucose-Starved Bacillus subtilis. J. Bacteriol. 2007, 190, 321–331.
- Frees, D.; Gerth, U.; Ingmer, H. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int. J. Med. Microbiol. 2014, 304, 142–149.
- Ujiie, H.; Matsutani, T.; Tomatsu, H.; Fujihara, A.; Ushida, C.; Miwa, Y.; Fujita, Y.; Himeno, H.; Muto, A. Trans-Translation is Involved in the CcpA-Dependent Tagging and Degradation of TreP in Bacillus subtilis. J. Biochem. 2008, 145, 59–66.
- Sauer, R.T.; Baker, T.A. AAA Proteases: ATP-Fueled Machines of Protein Destruction. Annu. Rev. Biochem. 2011, 80, 587–612.
- Schlothauer, T.; Mogk, A.; Dougan, D.A.; Bukau, B.; Turgay, K. MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. USA 2003, 100, 2306–2311.
- Turgay, K.; Hahn, J.; Burghoorn, J.; Dubnau, D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998, 17, 6730–6738.
- Pan, Q.; Garsin, D.A.; Losick, R. Self-Reinforcing Activation of a Cell-Specific Transcription Factor by Proteolysis of an Anti-σ Factor in B. subtilis. Mol. Cell 2001, 8, 873–883.
- Nakano, S.; Zheng, G.; Nakano, M.M.; Zuber, P. Multiple Pathways of Spx (YjbD) Proteolysis in Bacillus subtilis. J. Bacteriol. 2002, 184, 3664–3670.
- Miethke, M.; Hecker, M.; Gerth, U. Involvement of Bacillus subtilis ClpE in CtsR Degradation and Protein Quality Control. J. Bacteriol. 2006, 188, 4610–4619.
- Benaroudj, N.; Raynal, B.; Miot, M.; Ortiz-Lombardia, M. Assembly and proteolytic processing of mycobacterial ClpP1 and ClpP2. BMC Biochem. 2011, 12, 61.
- Donegan, N.P.; Marvin, J.S.; Cheung, A.L. Role of adaptor TrfA and ClpPC in controlling levels of SsrA-tagged proteins and antitoxins in Staphylococcus aureus. J. Bacteriol. 2014, 196, 4140–4151.
- Kirstein, J.; Schlothauer, T.; Dougan, D.A.; Lilie, H.; Tischendorf, G.; Mogk, A.; Bukau, B.; Turgay, K. Adaptor protein controlled oligomerization activates the AAA protein ClpC. EMBO J. 2006, 25, 1481–1491.
- Famulla, K.; Sass, P.; Malik, I.; Akopian, T.; Kandror, O.; Alber, M.; Hinzen, B.; Ruebsamen-Schaeff, H.; Kalscheuer, R.; Goldberg, A.L.; et al. Acyldepsipeptide antibiotics kill mycobacteria by preventing the physiological functions of the ClpP1P2 protease. Mol. Microbiol. 2016, 101, 194–209.
- Leodolter, J.; Warweg, J.; Weber-Ban, E. The Mycobacterium tuberculosis ClpP1P2 Protease Interacts Asymmetrically with Its ATPase Partners ClpX and ClpC1. PLoS ONE 2015, 10, e0125345.
- Schmitz, K.R.; Sauer, R.T. Substrate delivery by the AAA ClpX and ClpC1 unfoldases activates the mycobacterial ClpP1P2 peptidase. Mol. Microbiol. 2014, 93, 617–628.
- Zeiler, E.; List, A.; Alte, F.; Gersch, M.; Wachtel, R.; Poreba, M.; Drag, M.; Groll, M.; Sieber, S.A. Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad. Proc. Natl. Acad. Sci. USA 2013, 110, 11302–11307.
- Dahmen, M.; Vielberg, M.; Groll, M.; Sieber, S.A. Structure and Mechanism of the Caseinolytic Protease ClpP1/2 Heterocomplex from Listeria monocytogenes. Angew. Chem. Int. Ed. 2015, 54, 3598–3602.
- Pan, S.; Malik, I.T.; Thomy, D.; Henrichfreise, B.; Sass, P. The functional ClpXP protease of Chlamydia trachomatis requires distinct clpP genes from separate genetic loci. Sci. Rep. 2019, 9, 14129.
- Sekulovic, O.; Fortier, L. Global Transcriptional Response of Clostridium difficile Carrying the ϕCD38-2 Prophage. Appl. Environ. Microbiol. 2014, 81, 1364–1374.
