Retinal vascular disease is a highly prevalent vision-threatening ocular disease in the global population; however, its exact mechanism remains unclear. The expansion of omics technologies has revolutionized a new medical research methodology that combines multiple omics data derived from the same patients to generate multi-dimensional and multi-evidence-supported holistic inferences, providing unprecedented opportunities to elucidate the information flow of complex multi-factorial diseases. Omics data from eye biopsies can identify the molecular mechanisms of retinal vascular diseases in addition to the associated diagnostic prognostic and predictive biomarkers.
- multi-omics
- retinal vascular diseases
- biomarker
- therapeutic strategies
1. Current Potential Biomarkers for the Screening, Diagnosis, and Prognosis of Retinal Vascular Diseases
2. Omics-Based Biomarkers Discoveries
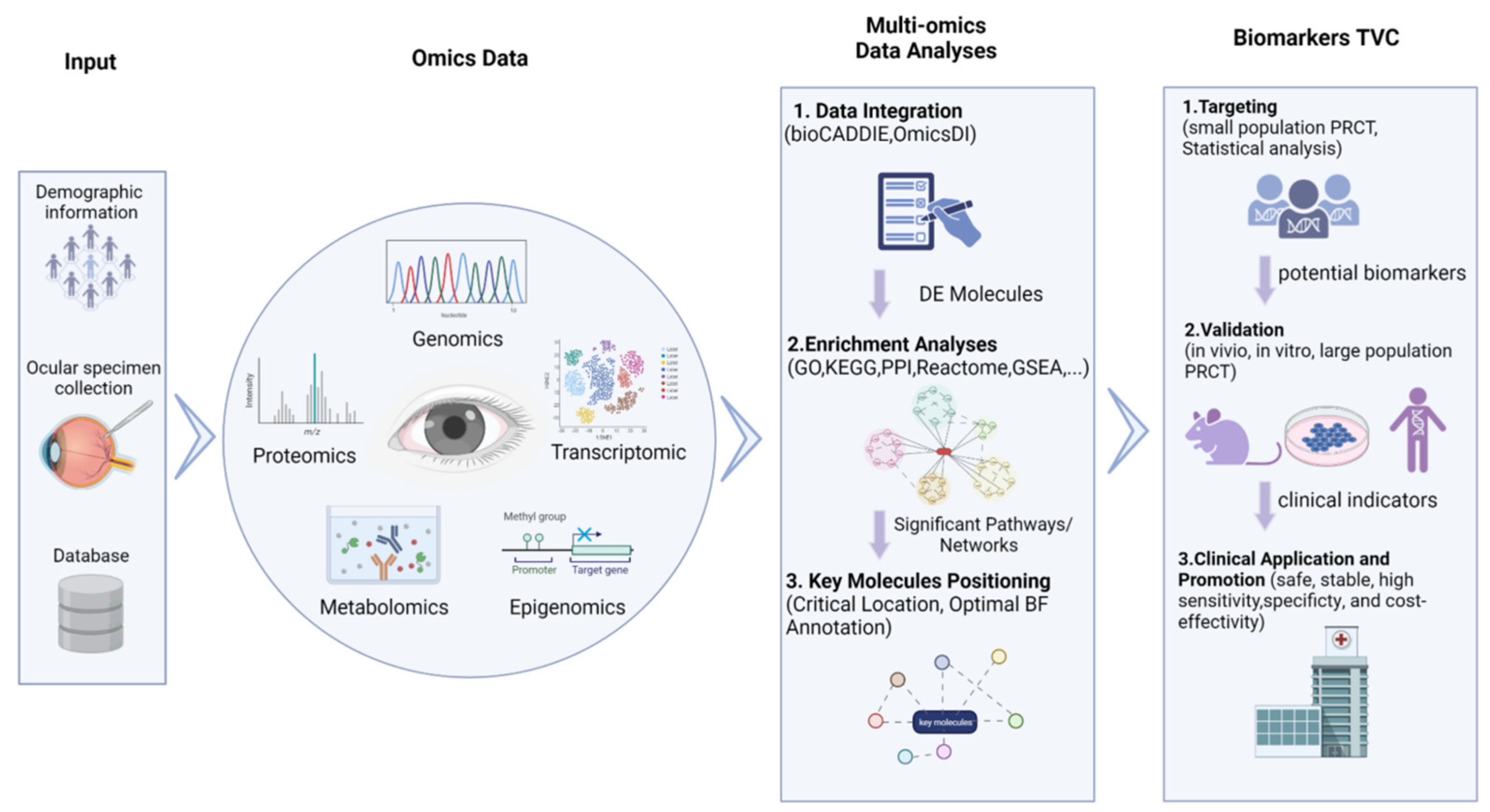
2.1. Source of Omics Data
2.2. Multi-Omics Data Integration Analysis
2.3. Biomarker Targeting, Validation, and Clinical Detection
3. Omics Biomarkers of Retinal Vascular Diseases
3.1. Omics Biomarkers in DR
3.2. Omics Biomarkers in AMD
3.3. Omics Biomarkers in RVO and ROP
3.4. Opportunities and Challenges That Coexist in Multi-Omics Integration Approaches Related to Biomarker Discoveries
This entry is adapted from the peer-reviewed paper 10.3390/cells12010103
References
- Gower, E.W.; Silverman, E.; Cassard, S.D.; Williams, S.K.; Baldonado, K.; Friedman, D.S. Barriers to attending an eye examination after vision screening referral within a vulnerable population. J. Health Care Poor Underserved 2013, 24, 1042–1052.
- Chin, E.K.; Ventura, B.V.; See, K.Y.; Seibles, J.; Park, S.S. Nonmydriatic fundus photography for teleophthalmology diabetic retinopathy screening in rural and urban clinics. Telemed. J. E Health 2014, 20, 102–108.
- Jiang, Z.; Yu, Z.; Feng, S.; Huang, Z.; Peng, Y.; Guo, J.; Ren, Q.; Lu, Y. A super-resolution method-based pipeline for fundus fluorescein angiography imaging. Biomed. Eng. Online 2018, 17, 125.
- Mesquita, J.; Castro-de-Sousa, J.P.; Vaz-Pereira, S.; Neves, A.; Passarinha, L.A.; Tomaz, C.T. Vascular endothelial growth factors and placenta growth factor in retinal vasculopathies: Current research and future perspectives. Cytokine Growth Factor Rev. 2018, 39, 102–115.
- Sivaprasad, S.; Prevost, T.; Bainbridge, J.; Edwards, R.T.; Hopkins, D.; Kelly, J.; Luthert, P.; Murphy, C.; Ramu, J.; Sarafraz-Shekary, N.; et al. Clinical efficacy and mechanistic evaluation of aflibercept for proliferative diabetic retinopathy (acronym CLARITY): A multicentre phase IIb randomised active-controlled clinical trial. BMJ Open 2015, 5, e008405.
- Zehetner, C.; Bechrakis, N.E.; Stattin, M.; Kirchmair, R.; Ulmer, H.; Kralinger, M.T.; Kieselbach, G.F. Systemic counterregulatory response of placental growth factor levels to intravitreal aflibercept therapy. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3279–3286.
- Nomura, Y.; Kaneko, M.; Miyata, K.; Yatomi, Y.; Yanagi, Y. Bevacizumab and Aflibercept Activate Platelets via FcγRIIa. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8075–8082.
- Loukovaara, S.; Nurkkala, H.; Tamene, F.; Gucciardo, E.; Liu, X.; Repo, P.; Lehti, K.; Varjosalo, M. Quantitative Proteomics Analysis of Vitreous Humor from Diabetic Retinopathy Patients. J. Proteome Res. 2015, 14, 5131–5143.
- Sun, J.; Glassman, A.R.; Beaulieu, W.T.; Stockdale, C.R.; Bressler, N.M.; Flaxel, C.; Gross, J.G.; Shami, M.; Jampol, L.M. Rationale and Application of the Protocol S Anti-Vascular Endothelial Growth Factor Algorithm for Proliferative Diabetic Retinopathy. Ophthalmology 2019, 126, 87–95.
- Kaštelan, S.; Orešković, I.; Bišćan, F.; Kaštelan, H.; Gverović Antunica, A. Inflammatory and angiogenic biomarkers in diabetic retinopathy. Biochem. Med. 2020, 30, 030502.
- Ting, D.S.; Tan, K.A.; Phua, V.; Tan, G.S.; Wong, C.W.; Wong, T.Y. Biomarkers of Diabetic Retinopathy. Curr. Diab. Rep. 2016, 16, 125.
- Lambert, N.G.; ElShelmani, H.; Singh, M.K.; Mansergh, F.C.; Wride, M.A.; Padilla, M.; Keegan, D.; Hogg, R.E.; Ambati, B.K. Risk factors and biomarkers of age-related macular degeneration. Prog. Retin. Eye Res. 2016, 54, 64–102.
- Wang, B.; Zhang, X.; Chen, H.; Koh, A.; Zhao, C.; Chen, Y. A Review of Intraocular Biomolecules in Retinal Vein Occlusion: Toward Potential Biomarkers for Companion Diagnostics. Front. Pharmacol. 2022, 13, 859951.
- Hartnett, M.E. Role of cytokines and treatment algorithms in retinopathy of prematurity. Curr. Opin. Ophthalmol. 2017, 28, 282–288.
- Panis, C.; Scandolara, T.B.; Pires, B.R.B.; Vacario, B.; de Amorim, I.S.S.; Siqueira, P.B.; Serpeloni, J.M.; Mencalha, A.L.; Bonvicino, C.R. An Overview Regarding Pharmacogenomics and Biomarkers Discovery: Focus on Breast Cancer. Curr. Top. Med. Chem. 2022, 22, 1654–1673.
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729.
- Zhu, L.; Huang, Q.; Li, X.; Jin, B.; Ding, Y.; Chou, C.J.; Su, K.-J.; Zhang, Y.; Chen, X.; Hwa, K.Y.; et al. Serological Phenotyping Analysis Uncovers a Unique Metabolomic Pattern Associated With Early Onset of Type 2 Diabetes Mellitus. Front. Mol. Biosci. 2022, 9, 841209.
- Ladapo, J.A.; Budoff, M.J.; Sharp, D.; Kuo, J.Z.; Huang, L.; Maniet, B.; Herman, L.; Monane, M. Utility of a Precision Medicine Test in Elderly Adults with Symptoms Suggestive of Coronary Artery Disease. J. Am. Geriatr. Soc. 2018, 66, 309–315.
- Peña-Bautista, C.; Baquero, M.; Vento, M.; Cháfer-Pericás, C. Omics-based Biomarkers for the Early Alzheimer Disease Diagnosis and Reliable Therapeutic Targets Development. Curr. Neuropharmacol. 2019, 17, 630–647.
- Jiang, S.; Hinchliffe, T.E.; Wu, T. Biomarkers of an Autoimmune Skin Disease—Psoriasis. Genom. Proteom. Bioinform. 2015, 13, 224–233.
- Rantamäki, A.H.; Seppänen-Laakso, T.; Oresic, M.; Jauhiainen, M.; Holopainen, J.M. Human tear fluid lipidome: From composition to function. PLoS ONE 2011, 6, e19553.
- Ghosh, A.; Nishtala, K. Biofluid lipidome: A source for potential diagnostic biomarkers. Clin. Transl. Med. 2017, 6, 22.
- Jones, G.; Lee, T.J.; Glass, J.; Rountree, G.; Ulrich, L.; Estes, A.; Sezer, M.; Zhi, W.; Sharma, S.; Sharma, A. Comparison of Different Mass Spectrometry Workflows for the Proteomic Analysis of Tear Fluid. Int. J. Mol. Sci. 2022, 23, 2307.
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy 2021, 17, 1–382.
- Sansone, S.-A.; Gonzalez-Beltran, A.; Rocca-Serra, P.; Alter, G.; Grethe, J.S.; Xu, H.; Fore, I.M.; Lyle, J.; Gururaj, A.E.; Chen, X.; et al. DATS, the data tag suite to enable discoverability of datasets. Sci. Data 2017, 4, 170059.
- Perez-Riverol, Y.; Bai, M.; da Veiga LePrevost, F.; Squizzato, S.; Park, Y.M.; Haug, K.; Carroll, A.J.; Spalding, D.; Paschall, J.; Wang, M.; et al. Discovering and linking public omics data sets using the Omics Discovery Index. Nat. Biotechnol. 2017, 35, 406–409.
- Guo, C.; Jiang, D.; Xu, Y.; Peng, F.; Zhao, S.; Li, H.; Jin, D.; Xu, X.; Xia, Z.; Che, M.; et al. High-Coverage Serum Metabolomics Reveals Metabolic Pathway Dysregulation in Diabetic Retinopathy: A Propensity Score-Matched Study. Front. Mol. Biosci. 2022, 9, 822647.
- Li, T.; Xu, Y.; Shi, Y.; Chen, J.; Lin, S.; Zhu, J.; Xu, X.; Lu, L.; Zou, H. Genome-wide analysis of DNA methylation identifies S100A13 as an epigenetic biomarker in individuals with chronic (≥30 years) type 2 diabetes without diabetic retinopathy. Clin. Epigenetics 2020, 12, 77.
- Skol, A.D.; Jung, S.C.; Sokovic, A.M.; Chen, S.; Fazal, S.; Sosina, O.; Borkar, P.P.; Lin, A.; Sverdlov, M.; Cao, D.; et al. Integration of genomics and transcriptomics predicts diabetic retinopathy susceptibility genes. eLife 2020, 9, e59980.
- Curovic, V.R.; Suvitaival, T.; Mattila, I.; Ahonen, L.; Trošt, K.; Theilade, S.; Hansen, T.W.; Legido-Quigley, C.; Rossing, P. Circulating Metabolites and Lipids Are Associated to Diabetic Retinopathy in Individuals With Type 1 Diabetes. Diabetes 2020, 69, 2217–2226.
- Amorim, M.; Martins, B.; Caramelo, F.; Gonçalves, C.; Trindade, G.; Simão, J.; Barreto, P.; Marques, I.; Leal, E.C.; Carvalho, E.; et al. Putative Biomarkers in Tears for Diabetic Retinopathy Diagnosis. Front. Med. 2022, 9, 873483.
- Mammadzada, P.; Bayle, J.; Gudmundsson, J.; Kvanta, A.; André, H. Identification of Diagnostic and Prognostic microRNAs for Recurrent Vitreous Hemorrhage in Patients with Proliferative Diabetic Retinopathy. J. Clin. Med. 2019, 8, 2217.
- Brunmair, J.; Bileck, A.; Schmidl, D.; Hagn, G.; Meier-Menches, S.M.; Hommer, N.; Schlatter, A.; Gerner, C.; Garhöfer, G. Metabolic phenotyping of tear fluid as a prognostic tool for personalised medicine exemplified by T2DM patients. EPMA J. 2022, 13, 107–123.
- Li, X.; Cai, S.; He, Z.; Reilly, J.; Zeng, Z.; Strang, N.; Shu, X. Metabolomics in Retinal Diseases: An Update. Biology 2021, 10, 944.
- Jian, Q.; Wu, Y.; Zhang, F. Metabolomics in Diabetic Retinopathy: From Potential Biomarkers to Molecular Basis of Oxidative Stress. Cells 2022, 11, 3005.
- Hou, X.W.; Wang, Y.; Pan, C.W. Metabolomics in Diabetic Retinopathy: A Systematic Review. Investig. Ophthalmol. Vis. Sci. 2021, 62, 4.
- Afarid, M.; Mohsenipoor, N.; Parsaei, H.; Amirmoezzi, Y.; Ghofrani-Jahromi, M.; Jafari, P.; Mohsenipour, A.; Sanie-Jahromi, F. Assessment of macular findings by OCT angiography in patients without clinical signs of diabetic retinopathy: Radiomics features for early screening of diabetic retinopathy. BMC Ophthalmol. 2022, 22, 281.
- Lee, H.; Lee, M.; Chung, H.; Kim, H.C. Quantification of retinal vessel tortuosity in diabetic retinopathy using optical coherence tomography angiography. Retina 2018, 38, 976–985.
- Balaratnasingam, C.; Inoue, M.; Ahn, S.; McCann, J.; Dhrami-Gavazi, E.; Yannuzzi, L.A.; Freund, K.B. Visual Acuity Is Correlated with the Area of the Foveal Avascular Zone in Diabetic Retinopathy and Retinal Vein Occlusion. Ophthalmology 2016, 123, 2352–2367.
- Rasti, R.; Allingham, M.J.; Mettu, P.S.; Kavusi, S.; Govind, K.; Cousins, S.W.; Farsiu, S. Deep learning-based single-shot prediction of differential effects of anti-VEGF treatment in patients with diabetic macular edema. Biomed. Opt. Express 2020, 11, 1139–1152.
- Babiuch, A.S.; Wykoff, C.C.; Srivastava, S.K.; Talcott, K.; Zhou, B.; Hach, J.; Hu, M.; Reese, J.L.; Ehlers, J.P. Retinal leakage index dynamics on ultra-widefield fluorescein angiography in eyes treated with intravitreal aflibercept for proliferative diabetic retinopathy in the recovery study. Retina 2020, 40, 2175–2183.
- Kersten, E.; Paun, C.C.; Schellevis, R.L.; Hoyng, C.B.; Delcourt, C.; Lengyel, I.; Peto, T.; Ueffing, M.; Klaver, C.C.; Dammeier, S.; et al. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv. Ophthalmol. 2018, 63, 9–39.
- ElShelmani, H.; Brennan, I.; Kelly, D.J.; Keegan, D. Differential Circulating MicroRNA Expression in Age-Related Macular Degeneration. Int. J. Mol. Sci. 2021, 22, 12321.
- Chen, G.; Walmsley, S.; Cheung, G.C.M.; Chen, L.; Cheng, C.-Y.; Beuerman, R.W.; Wong, T.Y.; Zhou, L.; Choi, H. Customized Consensus Spectral Library Building for Untargeted Quantitative Metabolomics Analysis with Data Independent Acquisition Mass Spectrometry and MetaboDIA Workflow. Anal. Chem. 2017, 89, 4897–4906.
- Wei, L.; Liu, B.; Tuo, J.; Shen, D.; Chen, P.; Li, Z.; Liu, X.; Ni, J.; Dagur, P.; Sen, H.N.; et al. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Rep. 2012, 2, 1151–1158.
- Oliver, V.F.; Jaffe, A.E.; Song, J.; Wang, G.; Zhang, P.; Branham, K.E.; Swaroop, A.; Eberhart, C.G.; Zack, D.J.; Qian, J.; et al. Differential DNA methylation identified in the blood and retina of AMD patients. Epigenetics 2015, 10, 698–707.
- Schori, C.; Trachsel, C.; Grossmann, J.; Zygoula, I.; Barthelmes, D.; Grimm, C. The Proteomic Landscape in the Vitreous of Patients with Age-Related and Diabetic Retinal Disease. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD31–AMD40.
- Coronado, B.N.L.; da Cunha, F.B.S.; de Oliveira, R.M.; Nóbrega, O.D.T.; Ricart, C.A.O.; Fontes, W.; de Sousa, M.V.; de Ávila, M.P.; Martins, A.M.A. Novel Possible Protein Targets in Neovascular Age-Related Macular Degeneration: A Pilot Study Experiment. Front. Med. 2021, 8, 692272.
- Hou, X.W.; Wang, Y.; Pan, C.W. Metabolomics in Age-Related Macular Degeneration: A Systematic Review. Investig. Ophthalmol. Vis. Sci. 2020, 61, 13.
- Brown, C.N.; Green, B.D.; Thompson, R.B.; den Hollander, A.I.; Lengyel, I. Metabolomics and Age-Related Macular Degeneration. Metabolites 2018, 9, 4.
- van Leeuwen, E.M.; Emri, E.; Merle, B.M.; Colijn, J.M.; Kersten, E.; Cougnard-Gregoire, A.; Dammeier, S.; Meester-Smoor, M.; Pool, F.M.; de Jong, E.K.; et al. A new perspective on lipid research in age-related macular degeneration. Prog. Retin. Eye Res. 2018, 67, 56–86.
- Deng, Y.; Shuai, P.; Wang, H.; Zhang, S.; Li, J.; Du, M.; Huang, P.; Qu, C.; Huang, L. Untargeted metabolomics for uncovering plasma biological markers of wet age-related macular degeneration. Aging 2021, 13, 13968–14000.
- Liu, K.; Fang, J.; Jin, J.; Zhu, S.; Xu, X.; Xu, Y.; Ye, B.; Lin, S.-H.; Xu, X. Serum Metabolomics Reveals Personalized Metabolic Patterns for Macular Neovascular Disease Patient Stratification. J. Proteome Res. 2020, 19, 699–707.
- Song, P.; Xu, Y.; Zha, M.; Zhang, Y.; Rudan, I. Global epidemiology of retinal vein occlusion: A systematic review and meta-analysis of prevalence, incidence, and risk factors. J. Glob. Health 2019, 9, 010427.
- Yao, J.; Chen, Z.; Yang, Q.; Liu, X.; Chen, X.; Zhuang, M.; Liu, Q. Proteomic analysis of aqueous humor from patients with branch retinal vein occlusion-induced macular edema. Int. J. Mol. Med. 2013, 32, 1421–1434.
- Cehofski, L.J.; Kruse, A.; Kjærgaard, B.; Stensballe, A.; Honoré, B.; Vorum, H. Proteins involved in focal adhesion signaling pathways are differentially regulated in experimental branch retinal vein occlusion. Exp. Eye Res. 2015, 138, 87–95.
- Cehofski, L.J.; Kruse, A.; Alsing, A.N.; Sejergaard, B.F.; Nielsen, J.E.; Schlosser, A.; Sorensen, G.L.; Grauslund, J.; Honoré, B.; Vorum, H. Proteome Analysis of Aflibercept Intervention in Experimental Central Retinal Vein Occlusion. Molecules 2022, 27, 3360.
- Cehofski, L.J.; Kruse, A.; Alsing, A.N.; Sejergaard, B.F.; Nielsen, J.E.; Pedersen, S.; Muttuvelu, D.V.; Kirkeby, S.; Honoré, B.; Vorum, H. Aflibercept Intervention in Experimental Branch Retinal Vein Occlusion Results in Upregulation of DnaJ Homolog Subfamily C Member 17. J. Ophthalmol. 2021, 2021, 6690260.
- Xiong, X.; Chen, X.; Ma, H.; Zheng, Z.; Yang, Y.; Chen, Z.; Zhou, Z.; Pu, J.; Chen, Q.; Zheng, M. Metabolite Changes in the Aqueous Humor of Patients With Retinal Vein Occlusion Macular Edema: A Metabolomics Analysis. Front. Cell Dev. Biol. 2021, 9, 762500.
- Fierson, W.M. Screening Examination of Premature Infants for Retinopathy of Prematurity. Pediatrics 2018, 142, e20183061.
- Good, W.V.; Gendron, R.L. Genomics and proteomics of retinopathy of prematurity: DNA-based prevention and treatment. Br. J. Ophthalmol. 2007, 91, 1577.
- Danielsson, H.; Tebani, A.; Zhong, W.; Fagerberg, L.; Brusselaers, N.; Hård, A.-L.; Uhlén, M.; Hellström, A. Blood protein profiles related to preterm birth and retinopathy of prematurity. Pediatr. Res. 2022, 91, 937–946.
- Xu, M.; Jiang, Y.; Su, L.; Chen, X.; Shao, X.; Ea, V.; Shang, Z.; Zhang, X.; Barnstable, C.J.; Li, X.; et al. Novel Regulators of Retina Neovascularization: A Proteomics Approach. J. Proteome Res. 2022, 21, 101–117.
- Zhou, Y.; Xu, Y.; Zhang, X.; Zhao, P.; Gong, X.; He, M.; Cao, J.; Jiang, B.; Yoshida, S.; Li, Y. Plasma metabolites in treatment-requiring retinopathy of prematurity: Potential biomarkers identified by metabolomics. Exp. Eye Res. 2020, 199, 108198.
- Yang, Y.; Wu, Z.; Li, S.; Yang, M.; Xiao, X.; Lian, C.; Wen, W.; He, H.; Zeng, J.; Wang, J.; et al. Targeted Blood Metabolomic Study on Retinopathy of Prematurity. Investig. Ophthalmol. Vis. Sci. 2020, 61, 12.
