Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Cell Biology
Mitochondria abundance and activity are fundamental elements underlying the dynamic adaptation of cells to stress conditions. The shape and number of mitochondria are tightly controlled by key biological processes, such as fusion and fission (also known as mitochondrial dynamics) and mitophagy that operate in an interconnected and dynamic way to sustain cellular health and metabolic needs.
- UPS
- mitochondria
- cancer
- dynamics
- metabolism
1. Mitochondria Dynamics
Mitochondrial fusion and fission are reversible rearrangements of mitochondrial structure. Mitochondria can fuse or divide their inner and outer membranes, assuming an elongated shape or punctiform pattern and promoting the cellular adaptation to changes in nutrients and oxygen availability, to cope cellular stress conditions. Mitochondria fusion relies on the concerted action of two distinct GTPase family proteins, mitofusins (MFN1/2) and optic atrophy 1 (OPA1). MFN1/2 are two GTPases that mediate the OMM fusion, whereas the fusion of inner mitochondrial membranes is mediated by cristae remodeling protein OPA-1 [1]. On the other hand, the mitochondrial fission process depends on the activity of dynamin-related protein 1 (DRP1) that is recruited on the organelle by direct binding to Fis1, an integral protein located at the OMM [2][3]. This binding allows the oligomerization of DRP1 that forms a ring structure around the mitochondria, constricting the mitochondrial outer membrane and causing the separation of the divided organelles [1]. In the yeast, the E3 ligase MDM30 ubiquitinates and degrades the mitofusin Fzo1p, thus inhibiting the mitochondrial fusion [4]. This process is finely counter regulated by two distinct ubiquitin binding proteins, UBP12 and UBP2, that were originally reported as DUBs that negatively regulate plant immunity [5]. Both DUBs are involved in the removal of ubiquitin moieties from different lysine residues of Fzo1p, with opposite effects on mitochondrial dynamics. In particular, UBP12 removes ubiquitin moieties and stabilizes Fzo1p, while UBP2 removes ubiquitin moieties and directs Fzo1p to the proteasome. Accordingly, cells lacking UBP2 show fragmented and smaller mitochondria, due to the impairment of the fission machinery, while cells lacking UBP12 show increased mitochondrial fusion [6]. In mammals, mitochondrial dynamics is tightly regulated by the UPS (Figure 1). Thus, the E3 ligase MUL1 localized at OMM, coordinates the ubiquitination and proteolysis of MFN2, limiting the mitochondrial fusion [7]. Furthermore, MUL1 sumoylates and stabilizes DRP1, preventing its proteolysis and promoting mitochondrial fission [8] and apoptosis [9]. These two important functions make mitochondrial MUL1 a key player in mitochondrial dynamics with a deep impact on cell survival. The fission mechanism is also regulated by the UPS. Thus, DRP1 and hFIS1 are ubiquitinated and degraded by the OMM E3 ligase MARCH5/MITOL, preventing mitochondrial fission [10]. Moreover, the E3 ligase synviolin (SYVN1) ubiquitinates and directs DRP1 to proteasome in the human ovarian granulosa-like tumor cell line (KGN). SYVN1 levels are markedly reduced in primary granulosa cells derived from polycystic ovary syndrome patients, resulting in increased DRP1 levels and mitochondrial fission [11].
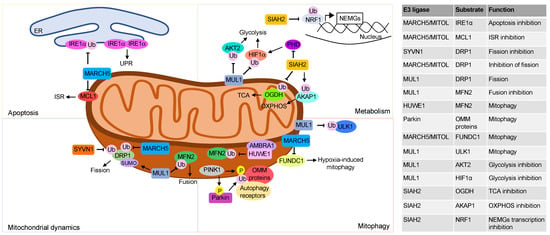
Figure 1. Mitochondrial regulation by UPS. The top left panel describes the ubiquitination events involving mitochondria E3 ligases that regulate apoptosis. The mitochondria E3 ligase MARCH5 ubiquitinates IRE1α located at the mitochondria associated membranes (MAMs) and inhibits apoptosis. Activation of ISR by anti-tumor drugs in solid tumors induces MARCH5-mediated ubiquitination and proteolysis of the antiapoptotic protein MCL1. The bottom left panel shows how the UPS regulates mitochondria dynamics, principally through post-translation modifications of DRP1 and MFN2. DRP1 sumoylation by MUL1 stabilizes the protein and promotes mitochondrial fission. Ubiquitin-dependent proteolysis of DRP1 by MARCH5 and SYVN1 inhibits mitochondrial fission. Moreover, the E3 ligase MUL1 by ubiquitinating and degrading MFN2 inhibits mitochondrial fusion. The top right panel describes ubiquitination events that regulate mitochondrial metabolism. The mitochondrial E3 ligase MUL1 ubiquitinates and degrades the cytoplasmatic proteins AKT2 and HIF1α rewiring cellular metabolism. The E3 ligase SIAH2 ubiquitinates and degrades the OGDH subunit of α ketoglutarate dehydrogenase, switching the glutamine metabolism to fatty acid synthesis. Moreover SIAH2, under hypoxia conditions, ubiquitinates and degrades AKAP1, reducing oxidative phosphorylation. Finally, SIAH2 ubiquitinates the transcription factor NRF1 and inhibits the transcription of the nuclear encoded mitochondrial genes (NEMGs). The bottom right panel shows how UPS at mitochondrial compartment regulates mitophagy. In response to mitochondrial damage, PINK1 accumulates at the OMM, phosphorylates ubiquitin, and recruits and activates Parkin. Parkin ubiquitinates different proteins at the outer mitochondrial membrane that are, then, recognized by autophagy receptors that promote mitophagy. Following oxidative stress, AMBRA1 recruits the E3 ligase HUWE1 at OMM. Here, HUWE1 ubiquitinates and degrades MFN2, favoring the elimination of damaged mitochondria. The mitochondrial E3 ligase MUL1 ubiquitinates ULK1 and promotes mitophagy. MARCH5 binds to- and ubiquitinates FUNDC1, promoting its proteasomal degradation. Downregulation of FUNDC1 regulates hypoxia-induced mitophagy.
2. Mitochondrial Quality Control
The number of mitochondria is tightly regulated through two different processes that promote the formation (biogenesis) or the elimination (mitophagy) of mitochondria. The mitochondrial biogenesis is an evolutionary conserved mechanism of self-replication through which cells enhance the number of mitochondria in response to cellular stresses, such as oxidative stress, or as consequence of extracellular stimulation by hormones, growth factors or endurance training [12]. A coordinated action of mitochondrial genome replication, transcription and translation of mitochondrial and nuclear genes supports the synthesis of components of the oxidative phosphorylation machinery, tricarboxylic acid cycle, mitochondrial metabolic pathways and membranes, promoting the formation of new organelles. Depending on specific metabolic needs and environmental conditions, the number of mitochondria varies in a cell-type dependent context [13].
Mitophagy is a conserved cellular mechanism that eliminates damaged or unneeded mitochondria through the autophagy pathway, avoiding the accumulation of dysfunctional organelles [14]. It is well known that oxidative stress regulates the mitophagy pathway through UPS. In response to mitochondrial damage or hypoxia, the mitochondrial membrane potential decreases, causing the stabilization and accumulation of the PTEN induced kinase 1 (PINK1) protein at the OMM [15]. The cytosolic E3 ubiquitin ligase Parkin, then, is rapidly recruited on mitochondria by PINK1. Once at the OMM, Parkin ubiquitinates different mitochondrial targets including the voltage dependent anion-selective channel (VDAC) and mitofusins 1/2 (MFN1/2). The ubiquitin moieties bound to these substrates furnish docking sites for autophagy receptors, allowing the initiation of the mitophagy process and the consequent removal of damaged mitochondria from the cell [16]. Although the PINK1/Parkin axis is the best characterized mechanism of mitophagy control exerted by the UPS, mitophagy can occur also in cells lacking Parkin, indicating the presence of additional players and mechanisms involved in the process. Accordingly, in the last few years, other E3 ligases have been connected with the initiation of mitophagy process in response to oxidative damage. In particular, the activating molecule in Beclin 1-regulated autophagy protein 1 (AMBRA1), the cytosolic HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 (HUWE1) and MARCH5 have been connected with connected with Parkin-independent mitophagy. Thus, in response to oxidative stress, AMBRA1 binds and recruits HUWE1 to the OMM. HUWE1, in turn, ubiquitinates and directs MFN2 to proteasome, impairing the fusion of damaged mitochondria and favoring their elimination through mitophagy [17][18]. The mitochondrial E3 ligase MARCH5 regulates hypoxia-induced mitophagy by ubiquitinating and degrading the FUN14 domain containing protein 1 (FUNDC1). Proteolysis of FUNDC1 desensitizes mitochondria to hypoxia-induced mitophagy [19].
This entry is adapted from the peer-reviewed paper 10.3390/cells12020234
References
- Senft, D.; Ronai, Z.A. Regulators of mitochondrial dynamics in cancer. Curr. Opin. Cell Biol. 2016, 39, 43–52.
- Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010, 11, 872–884.
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159.
- Cohen, M.M.J.; Leboucher, G.P.; Livnat-Levanon, N.; Glickman, M.H.; Weissman, A.M. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell 2008, 19, 2457–2464.
- Ewan, R.; Pangestuti, R.; Thornber, S.; Craig, A.; Carr, C.; O’Donnell, L.; Zhang, C.; Sadanandom, A. Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 2011, 191, 92–106.
- Anton, F.; Dittmar, G.; Langer, T.; Escobar-Henriques, M. Two Deubiquitylases Act on Mitofusin and Regulate Mitochondrial Fusion along Independent Pathways. Mol. Cell 2013, 49, 487–498.
- Yun, J.; Puri, R.; Yang, H.; Lizzio, M.A.; Wu, C.; Sheng, Z.H.; Guo, M. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife 2014, 3, e01958.
- Braschi, E.; Zunino, R.; McBride, H.M. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009, 10, 748–754.
- Prudent, J.; Zunino, R.; Sugiura, A.; Mattie, S.; Shore, G.C.; McBride, H.M. MAPL SUMOylation of Drp1 Stabilizes an ER/Mitochondrial Platform Required for Cell Death. Mol. Cell 2015, 59, 941–955.
- Yonashiro, R.; Ishido, S.; Kyo, S.; Fukuda, T.; Goto, E.; Matsuki, Y.; Ohmura-Hoshino, M.; Sada, K.; Hotta, H.; Yamamura, H.; et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006, 25, 3618–3626.
- Sun, L.; Ye, H.; Tian, H.; Xu, L.; Cai, J.; Zhang, C.; Wang, R.; Yang, H.; Zhao, S.; Zhang, J.; et al. The E3 Ubiquitin Ligase SYVN1 Plays an Antiapoptotic Role in Polycystic Ovary Syndrome by Regulating Mitochondrial Fission. Oxid. Med. Cell Longev. 2022, 2022, 3639302.
- Bennett, C.F.; Latorre-Muro, P.; Puigserver, P. Mechanisms of mitochondrial respiratory adaptation. Nat. Rev. Mol. Cell Biol. 2022, 23, 817–835.
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284.
- Harper, J.W.; Ordureau, A.; Heo, J.-M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 93–108.
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.-S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221.
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314.
- Di Rita, A.; Peschiaroli, A.; Strobbe, D.; Hu, Z.; Gruber, J.; Nygaard, M.; Lambrughi, M.; Melino, G.; Papaleo, E.; Dengjel, J.; et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 2018, 9, 3755.
- Strappazzon, F.; Di Rita, A.; Peschiaroli, A.; Leoncini, P.P.; Locatelli, F.; Melino, G.; Cecconi, F. HUWE1 controls MCL1 stability to unleash AMBRA1-induced mitophagy. Cell Death Differ. 2019, 27, 1155–1168.
- Chen, Z.; Liu, L.; Cheng, Q.; Li, Y.; Wu, H.; Zhang, W.; Wang, Y.; Sehgal, S.A.; Siraj, S.; Wang, X.; et al. Mitochondrial E3 ligase MARCH 5 regulates FUNDC 1 to fine-tune hypoxic mitophagy. EMBO Rep. 2017, 18, 495–509.
This entry is offline, you can click here to edit this entry!
