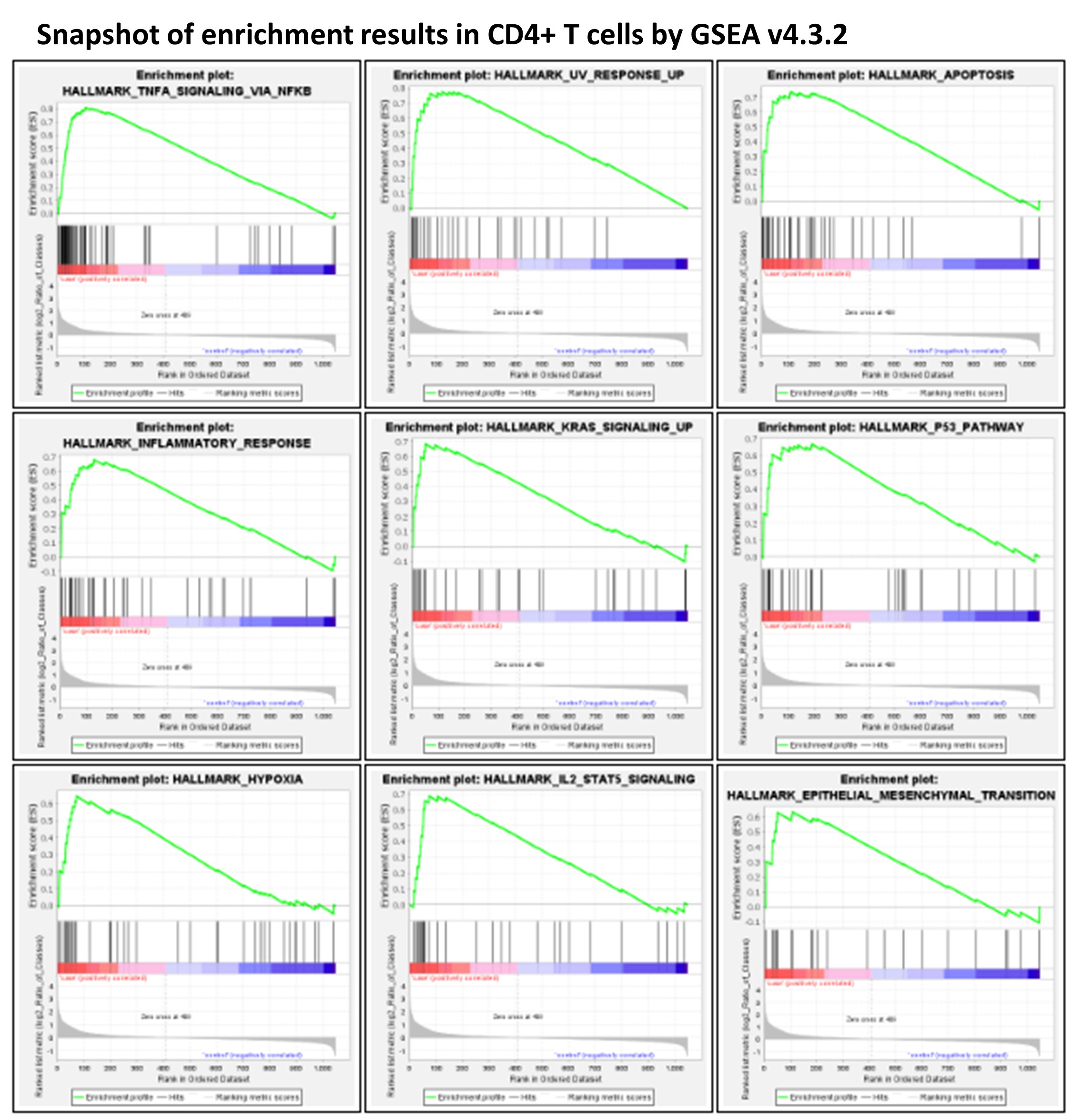
With highly sensitive single cell RNA-seq (scRNAseq), our study highlights the effects of blood storage on PBMCs, e.g. gene sets highly relevant to human diseases (NF-kB, AP-1 signaling), and advocates for scRNAseq/bulk RNAseq experiments of PBMCs from fresh blood.
- AP-1
- NF-kB
- peripheral blood mononuclear cells
- single cell RNA-seq
- transcriptome
Peripheral blood mononuclear cells (PBMCs) are widely used as a model in the study of different human diseases. There is often a time delay from blood collection to PBMC isolation during the sampling process, which can result in an experimental bias, particularly when performing highly sensitive single cell RNA-seq (scRNAseq) studies. This study examined the impact of different time periods from blood draw to PBMC isolation on the subsequent transcriptome profiling of different cell types in PBMCs by scRNAseq using the 10X Chromium Single Cell Gene Expression assay. Examining the five major cell types constituting the PBMC cell population, i.e., CD4+ T cells, CD8+ T cells, NK cells, monocytes, and B cells, both common changes and cell-type-specific changes were observed in the single cell transcriptome profiling over time. In particular, the upregulation of genes regulated by NF-kB in response to TNF was observed in all five cell types. Significant changes in key genes involved in AP-1 signaling were also observed. RBC contamination was a major issue in stored blood, whereas RBC adherence had no direct impact on the cell transcriptome. This study highlights the effects of blood storage on PBMCs and advocates for scRNAseq/bulk RNAseq experiments of PBMCs from fresh blood. Some major changes of transcriptome highly relevant to human diseases (e.g. NF-kB, AP-1 signaling) in stored blood as shown by this study may not be corrected by computational approaches. For more details, please see https://www.mdpi.com/2073-4425/14/1/142
This entry is adapted from the peer-reviewed paper 10.3390/genes14010142
