Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Sulfur is not only one of the most abundant elements on the Earth, but it is also essential to all living organisms. As life likely began and evolved in a hydrogen sulfide (H2S)-rich environment, sulfur metabolism represents an early form of energy generation via various reactions in prokaryotes and has driven the sulfur biogeochemical cycle since.
- reactive sulfur species
- hydrogen sulfide
- sulfur transformation
1. Introduction
Sulfur, one of the most abundant elements on our planet, and an element that has six valence electrons, is essential to all living organisms. The term ‘reactive sulfur species (RSS)′, coined by Jacob and colleagues [1], has been in the general scientific vocabulary for more than two decades. Apparently, RSS, much like reactive oxygen species (ROS) and reactive nitrogen species (RNS), comprise chemically reactive molecules containing the element for which they are named, sulfur, oxygen, and nitrogen respectively [2]. Because of their highly reactive nature, the reactive species are prone to interact with cellular macromolecules, including nucleic acids, proteins, and lipids [3]. Not surprisingly, most, if not all, of these reactive species, all of which can be generated endogenously in the cell and arise abiotically in environments, are hostile to living cells at supraphysiological levels [2]. Despite this, in eukaryotes, especially mammals, a large majority of studies in fact center on the beneficial roles that these species play by mediating redox signaling and redox regulation [4]. It has been firmly established that these reactive species, as a signaling molecules, impact diverse processes including metabolism, vasodilation, neurotransmission, immunity, apoptosis, and cancer [2][5][6].
Compared to ROS and RNS, which have been extensively studied for decades, RSS are relatively new. The concept of RSS has evolved over time and is still far from unanimously held. The ‘narrow’ view defines RSS as the molecules produced from sequential one-electron oxidations of hydrogen sulfide (H2S), forming a thiyl (sulfhydryl) radical (HS•, the counterpart of HO• in ROS, the same below), hydrogen persulfide (H2S2, H2O2), and the supersulfide radical (S2•, O2•) [7] (Figure 1). In the ‘broad’ view, RSS refer collectively to reactive sulfur chemotypes (both organic and inorganic) that can react with, oxidize or reduce other molecules under physiological conditions [8][9] (Figure 1). Therefore, ‘reactive’ sulfur atoms can be found in compounds having sulfur of higher valence, such as sulfite (SO32−, +4) and sulfate (SO42−, +6), which have interchalcogen bonds, and thiosulfate (S2O32−, −1/+5 or 0/+4), in which sulfur atoms are catenated as in polysulfides (Figure 1). To date, the widely accepted notion about RSS, which researchers defined here as the ‘normal’ view, regards RSS as any molecules carrying reactive sulfur with a valence of −2, −1, or 0 [9][10]. In this sense, representative RSS include protein thiols (PSH) and persulfide (PSSH), low-molecule-weight (LMW) thiols (RSH) and persulfide (RSSH), hydrogen persulfide/polysulfide (H2Sn, n ≥ 2), polysulfides (RSS (n) R, n > 1), and sulfenic acids (RSOH) [10][11] (Figure 1).
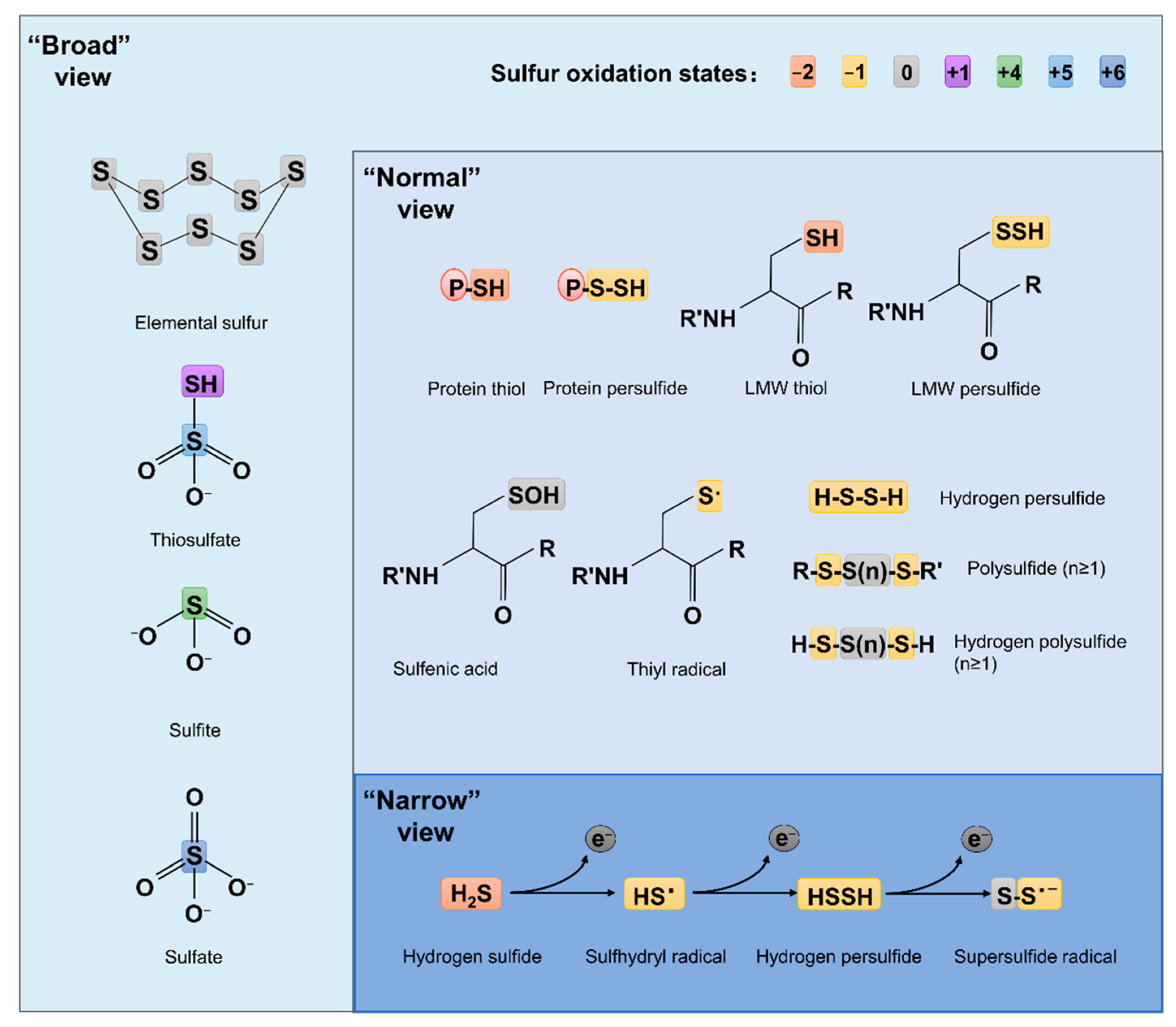
Figure 1. Structures of some RSS chemotypes in different views. The red (−2), yellow (−1), gray (0), purple (+1), green (+4), light blue (+5) and dark blue (+6) rectangles are used to designate the valence states of sulfur, as specified.
Formation and metabolism of RSSs have been intensively studied in eukaryotes, especially mammals [10][12]. In these processes, H2S is the center molecule that is generated in the cytosol through degradation of thiol-containing molecules, cysteine (cys) in particular [9] (Figure 2). Persulfide and polysulfide can then be formed from H2S through various non-enzymatic reactions involving ROS. While bacteria preserve these pathways to generate H2S, many of them can also do so through reduction of a variety of inorganic and organic compounds containing sulfur of high higher valence, releasing H2S as well as other RSS as the terminal products or intermediates [13][14]. In parallel, H2S can be oxidized by diverse prokaryotes, and even mitochondria, through a combination of abiotic and biotic reactions to diverse sulfur compounds of any possible valence, many of which are RSS [8][9]. As inorganic sulfur in the form of sulfate (SO42−) is extremely abundant in oceans, and metabolites in sulfur cycling are diverse, RSSs generated by microbes have profound impacts on all life and beyond [15].
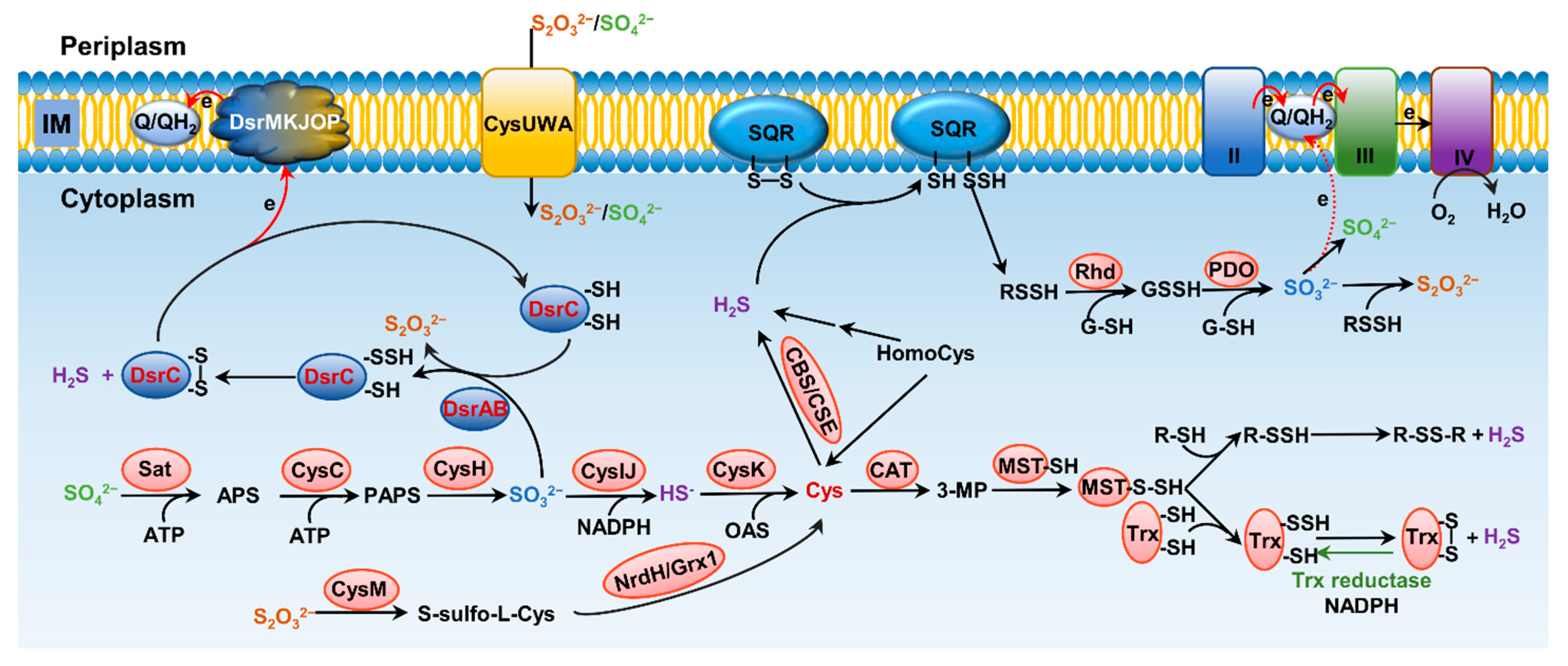
Figure 2. Pathways for bacterial sulfur metabolism in the cytoplasm. H2S biogenesis through amino acid metabolism: generation of H2S from homocys can be catalyzed by cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE). Cys aminotransferase (CAT) catalyzes the formation of 3-mercaptopyruvate (3-MP) from cys, and then, mercaptopyruvate sulfurtransferase (MST) converts 3-MP to H2S. H2S biogenesis occurs through assimilatory sulfate reduction (ASR) and dissimilatory sulfate reduction (DSR) of inorganic sulfur species; the latter only occurs in sulfate-reducing bacteria (SRB), catalyzing SO32− to HS− through dissimilatory sulfite reductase (Dsr). For ASR pathway, SO42−, which imported from ATP-dependent transporter CysUWA, is catalyzed and converted to HS− by a series of enzymes, including Sat, CysC, CysH, and CysIJ. S2O32− can also be reduced and used to synthesize cys by CysM and NrdH/Grx1. H2S catabolism: H2S binds to sulfur quinone oxidoreductase (SQR), and goes through a series of sequential reactions, including oxidation of sulfide to polysulfide by membrane-bound SQR, formation of GSSH from reaction involving with rhodanese (Rhd), oxidation of the sulfane sulfur in GSSH to SO32− catalyzed by persulfide dioxygenase (PDO), formation of S2O32− from spontaneous reaction between polysulfide and SO32−, and formation of SO42− either spontaneously or catalyzed by various enzymes, transferring two electrons via quinone into the electron transport chain.
Despite the difference in their functioning roles, the mechanism for biological activity of the reactive species is similar: modification of cellular targets. It is generally accepted that the reactive species interact with their specific cellular targets for redox signaling at physiological levels, whereas at elevated concentrations they modify macromolecules in a rather indiscriminate way, with interactions amounting to damaged enzymes and DNA, and even cell death [4]. This also holds true for RSS, although sulfur has been usually considered as the essential element constituting cellular antioxidant systems [16].
In recent years, a number of studies combining conventional genetic analysis and cutting-edge technologies have stressed the profound biological roles of RSS in mammalian as well as bacterial cells. While an understanding of the sensing, production, and physiological role of RSS in prokaryotes, bacteria in particular, is still in its infancy, it is clear that control of RSS homeostasis emerges as a promising means for therapeutic treatments for sulfur-related diseases, for maintenance of a balanced sulfur biogeochemical cycle, and for development of RSS-dependent biotechnologies.
2. Influence of Sulfur on the Origin of Life
Redox reactions, a result of electron imbalance mediated by energy, are the basis of all physiological activities in cells, such as respiration and photosynthesis. The original life began in an anoxic ferrous ocean 3.8 billion years ago (bya), likely as anoxygenic chemolithotrophs in environments associated with deep-sea hydrothermal vent systems [7][17][18]. As these environments were characterized by large amounts of sulfur and metals, iron in particular, it is believed that iron-sulfur clusters were the first electron-transfer units to be produced during chemical evolution [19]. In addition, sulfur now has been recognized to be a crucial element in the physiology of life’s common ancestor because some sulfur species, such as acetyl-coenzyme A, are essential intermediates in metabolism [20][21]. Moreover, H2S and diverse RSS, such as elemental sulfur, sulfite, thiosulfate, and polysulfides, rather than sulfate, which is the predominant inorganic sulfur compound on the current earth, were more prevalent in the ancient ocean and have played an critical role in shaping the sulfur cycle and life [22][23][24].
A large portion of contemporary living organisms, eukaryotes in particular, use O2 as the terminal acceptor for energy production. During oxygen respiration, ROS are generated endogenously in the cell. It has been postulated that antioxidant mechanisms, including superoxide dismutase, catalase, and peroxide reductase, evolved concomitantly with oxygen respiration [25]. However, the history of RSS is undoubtedly much longer than that of ROS, and more importantly, antioxidant mechanisms were present far earlier than the oxic ocean [26]. The anoxygenic photosynthesis appeared ~3–3.5 bya, several million years before the cyanobacterial existence, according to fossil evidence [27][28]. In the early photosynthetic organisms, their light-gathering antennae and processing machinery were not sophisticated enough to support oxidation of H2O, and instead, H2S was more likely to act as the electron donor [7][29][30]. From 2.3 billion years on, in the ‘great oxidation event (GOE)’, cyanobacteria had evolved ability to oxidize H2O and produced a large amount of oxygen via photosynthesis [31]. During that time, the atmospheric O2 fluctuated between 0%–2% for 1.7 billion years, while the oceans remained anoxic [17][29][30][32][33].
While the first eukaryotes appeared approximately 1.5 bya, it was not until 0.6 bya that the ocean finally became aerobic [25][34]. This time, called the ‘Boring Billion’, represents a period of geobiological stasis caused by prolonged nutrient, climate, atmospheric and tectonic stability [35]. Phylogenetic studies have shown that antioxidant enzymes were present in the last universal common ancestor, well before the emergence of cyanobacteria and the GOE [36]. These enzymes are present in all domains of life, even in anaerobic bacteria, suggesting that these enzymes are ancient and that the need for ROS protection is pervasive in non-aerobic environments [26][36][37][38][39][40][41][42]. Because of the chemical similarity between O and S, and the less electron-negativity and greater reactivity versatility of S, these enzymes have been proposed to be primarily used to resist RSS. In recent years, evidence supporting this proposal has emerged [26][43][44][45][46]. All of these insights have provided the new interpretation of RSS in biological evolution.
This entry is adapted from the peer-reviewed paper 10.3390/antiox11122487
References
- Giles, G.I.; Tasker, K.M.; Jacob, C. Hypothesis: The role of reactive sulfur species in oxidative stress. Free Radic. Biol. Med. 2001, 31, 1279–1283.
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383.
- Herb, M.; Schramm, M. Functions of ROS in macrophages and antimicrobial Immunity. Antioxidants 2021, 10, 313.
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709.
- Sies, H. Chapter 13—Oxidative Stress: Eustress and Distress in Redox Homeostasis. In Stress: Physiology, Biochemistry, and Pathology; Fink, G., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 153–163.
- Khattak, S.; Rauf, M.A.; Khan, N.H.; Zhang, Q.-Q.; Chen, H.-J.; Muhammad, P.; Ansari, M.A.; Alomary, M.N.; Jahangir, M.; Zhang, C.-Y.; et al. Hydrogen sulfide biology and its role in cancer. Molecules 2022, 27, 3389.
- Olson, K.R. Are reactive sulfur species the new reactive oxygen species? Antioxid. Redox Signal. 2020, 33, 1125–1142.
- Mishanina, T.V.; Libiad, M.; Banerjee, R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 2015, 11, 457–464.
- Filipovic, M.R.; Zivanovic, J.; Alvarez, B.; Banerjee, R. Chemical biology of H2S Signaling through persulfidation. Chem. Rev. 2018, 118, 1253–1337.
- Kolluru, G.K.; Shen, X.; Kevil, C.G. Reactive sulfur species. Arter. Thromb. Vasc. Biol. 2020, 40, 874–884.
- Lau, N.; Pluth, M.D. Reactive sulfur species (RSS): Persulfides, polysulfides, potential, and problems. Curr. Opin. Chem. Biol. 2019, 49, 1–8.
- Gruhlke, M.C.H.; Slusarenko, A.J. The biology of reactive sulfur species (RSS). Plant Physiol. Biochem. 2012, 59, 98–107.
- Moran, M.A.; Durham, B.P. Sulfur metabolites in the pelagic ocean. Nat. Rev. Microbiol. 2019, 17, 665–678.
- Yu, Q.; Sun, W.; Gao, H. Thiosulfate oxidation in sulfur-reducing Shewanella oneidensis and its unexpected influences on the cytochrome c content. Environ. Microbiol. 2021, 23, 7056–7072.
- Landa, M.; Burns, A.S.; Durham, B.P.; Esson, K.; Nowinski, B.; Sharma, S.; Vorobev, A.; Nielsen, T.; Kiene, R.P.; Moran, M.A. Sulfur metabolites that facilitate oceanic phytoplankton–bacteria carbon flux. ISME J. 2019, 13, 2536–2550.
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87.
- Olson, K.R. Reactive oxygen species or reactive sulfur species: Why we should consider the latter. J. Exp. Biol. 2020, 223, jeb196352.
- Sojo, V.; Herschy, B.; Whicher, A.; Camprubí, E.; Lane, N. The origin of life in alkaline hydrothermal vents. Astrobiology 2016, 16, 181–197.
- Garcia, P.S.; D’Angelo, F.; Ollagnier de Choudens, S.; Dussouchaud, M.; Bouveret, E.; Gribaldo, S.; Barras, F. An early origin of iron–sulfur cluster biosynthesis machineries before Earth oxygenation. Nat. Ecol. Evol. 2022, 6, 1564–1572.
- Nitschke, W.; Russell, M.J. Beating the acetyl coenzyme A-pathway to the origin of life. Philos. Trans. R. Soc. B 2013, 368, 20120258.
- Ritson, D.J. A cyanosulfidic origin of the Krebs cycle. Sci. Adv. 2021, 7, eabh3981.
- Wacey, D.; Kilburn, M.R.; Saunders, M.; Cliff, J.; Brasier, M.D. Microfossils of sulphur-metabolizing cells in 3.4-billion-year-old rocks of Western Australia. Nat. Geosci. 2011, 4, 698–702.
- Philippot, P.; Van Zuilen, M.; Lepot, K.; Thomazo, C.; Farquhar, J.; Van Kranendonk, M.J. Early archaean microorganisms preferred elemental sulfur, not sulfate. Science 2007, 317, 1534–1537.
- Marin, J.; Battistuzzi, F.U.; Brown, A.C.; Hedges, S.B. The timetree of prokaryotes: New insights into their evolution and speciation. Mol. Biol. Evol. 2016, 34, 437–446.
- Jones, D.P.; Sies, H. The redox code. Antioxid. Redox Signal. 2015, 23, 734–746.
- Olson, K.R.; Gao, Y.; DeLeon, E.R.; Arif, M.; Arif, F.; Arora, N.; Straub, K.D. Catalase as a sulfide-sulfur oxido-reductase: An ancient (and modern?) regulator of reactive sulfur species (RSS). Redox Biol. 2017, 12, 325–339.
- Schmidt, A. Sulfur metabolism in Cyanobacteria. Methods Enzymol. 1988, 167, 572–583.
- Schopf, J.W. Disparate rates, differing fates: Tempo and mode of evolution changed from the Precambrian to the Phanerozoic. Proc. Natl. Acad. Sci. USA 1994, 91, 6735–6742.
- Holland, H.D. Volcanic gases, black smokers, and the Great Oxidation Event. Geochim. Cosmochim. Acta 2002, 66, 3811–3826.
- Planavsky, N.J.; Bekker, A.; Hofmann, A.; Owens, J.D.; Lyons, T.W. Sulfur record of rising and falling marine oxygen and sulfate levels during the Lomagundi event. Proc. Natl. Acad. Sci. USA 2012, 109, 18300–18305.
- Poulton, S.W.; Bekker, A.; Cumming, V.M.; Zerkle, A.L.; Canfield, D.E.; Johnston, D.T. A 200-million-year delay in permanent atmospheric oxygenation. Nature 2021, 592, 232–236.
- Lyons, T.W.; Reinhard, C.T.; Planavsky, N.J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 2014, 506, 307–315.
- Schopf, J.W. Geological evidence of oxygenic photosynthesis and the biotic response to the 2400–2200 Ma “Great Oxidation Event”. Biochemistry 2014, 79, 165–177.
- Kurland, C.G.; Andersson, S.G.E. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 2000, 64, 786–820.
- Mukherjee, I.; Large, R.R.; Corkrey, R.; Danyushevsky, L.V. The Boring Billion, a slingshot for complex life on earth. Sci. Rep. 2018, 8, 4432.
- Case, A.J. On the origin of superoxide dismutase: An evolutionary perspective of superoxide-mediated redox signaling. Antioxidants 2017, 6, 82.
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349.
- Zamocky, M.; Gasselhuber, B.; Furtmuller, P.G.; Obinger, C. Molecular evolution of hydrogen peroxide degrading enzymes. Arch. Biochem. Biophys. 2012, 525, 131–144.
- Inupakutika, M.A.; Sengupta, S.; Devireddy, A.R.; Azad, R.K.; Mittler, R. The evolution of reactive oxygen species metabolism. J. Exp. Bot. 2016, 67, 5933–5943.
- Dibrova, D.V.; Cherepanov, D.A.; Galperin, M.Y.; Skulachev, V.P.; Mulkidjanian, A.Y. Evolution of cytochrome bc complexes: From membrane-anchored dehydrogenases of ancient bacteria to triggers of apoptosis in vertebrates. Biochim. Biophys. Acta-Bioenerg. 2013, 1827, 1407–1427.
- Grace, S.C. Phylogenetic distribution of superoxide-dismutase supports an endosymbiotic origin for chloroplasts and mitochondria. Life Sci. 1990, 47, 1875–1886.
- Miller, A.F. Superoxide dismutases: Ancient enzymes and new insights. Febs Lett. 2012, 586, 585–595.
- Hou, N.; Yan, Z.; Fan, K.; Li, H.; Zhao, R.; Xia, Y.; Xun, L.; Liu, H. OxyR senses sulfane sulfur and activates the genes for its removal in Escherichia coli. Redox Biol. 2019, 26, 101293.
- Sato, I.; Shimatani, K.; Fujita, K.; Abe, T.; Shimizu, M.; Fujii, T.; Hoshino, T.; Takaya, N. Glutathione reductase/glutathione is responsible for cytotoxic elemental sulfur tolerance via polysulfide shuttle in fungi. J. Biol. Chem. 2011, 286, 20283–20291.
- Dóka, É.; Pader, I.; Bíró, A.; Johansson, K.; Cheng, Q.; Ballagó, K.; Prigge, J.R.; Pastor-Flores, D.; Dick, T.P.; Schmidt, E.E.; et al. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2016, 2, e1500968.
- Liu, D.; Chen, J.; Wang, Y.; Meng, Y.; Li, Y.; Huang, R.; Xia, Y.; Liu, H.; Jiao, N.; Xun, L.; et al. Synechococcus sp. PCC7002 uses peroxiredoxin to cope with reactive sulfur species stress. mBio 2022, 13, e01039-22.
This entry is offline, you can click here to edit this entry!
