Inflammatory bowel disease (IBD), comprising Crohn’s disease (CD) and ulcerative colitis (UC), is characterized as a chronic and recurrent inflammatory disease whose pathogenesis is still elusive. The gut microbiota exerts important and diverse effects on host physiology through maintaining immune balance and generating health-benefiting metabolites. Many studies have demonstrated that IBD is associated with disturbances in the composition and function of the gut microbiota. Both the abundance and diversity of gut microbiota are dramatically decreased in IBD patients. Furthermore, some particular classes of microbiota-derived metabolites, principally short-chain fatty acids, tryptophan, and its metabolites, and bile acids have also been implicated in the pathogenesis of IBD.
- inflammatory bowel disease
- gut microbiota
- dysbiosis
- microbiota-derived metabolites
1. Introduction
Inflammatory bowel disease (IBD), generally including ulcerative colitis (UC) and Crohn’s disease (CD), is a chronic and relapsing intestinal inflammatory disease. There are differences in the predilection sites between UC and CD. UC usually involves the distal colon and rectum, invading the intestinal mucosa and submucosa, while CD, a discontinuous inflammation disease, can attack the entire digestive tract, and its lesions can penetrate all the intestinal layers [1]. IBD tends to occur in developed countries, and epidemiological surveys show that its incidence has gradually increased in recent years globally. It was reported that, from 1980 to 2013, the incidence of IBD in Denmark increased from 15.9 to 27.7 per 100,000 people [2]. Furthermore, the incidence of IBD in the Netherlands increased from 17.51 to 38.96 per 100,000 people from 1991 to 2010, with an average growth rate of 4.30% annually [3]. In recent years, the prevalence of IBD has increased in Asia, and it shows a gradient trend of increase from north to south in China [4]. Although the pathogenesis of IBD is still elusive, it is generally believed to be related to genetic susceptibility, environmental factors, immune dysfunction, and gut microbiota [5][6]. It was reported that a high risk of developing IBD was observed in the offspring of affected parents [7]. Potentially relevant environmental factors, such as antibiotic exposure, smoking, dietary fiber, saturated fats, and major life stressors, are associated with IBD incidence [8]. In addition, gut microbiota dysbiosis was demonstrated in IBD patients and consistently characterized by a reduction in microbiota diversity compared with healthy individuals [9]. The use of existing therapeutic drugs, including sulfasalazine, mesalazine, glucocorticoids, and biological drugs, is limited to some extent due to their modest effects, adverse reactions, and high prices [10]. Therefore, new treatment strategies and methods have been called for in recent years.
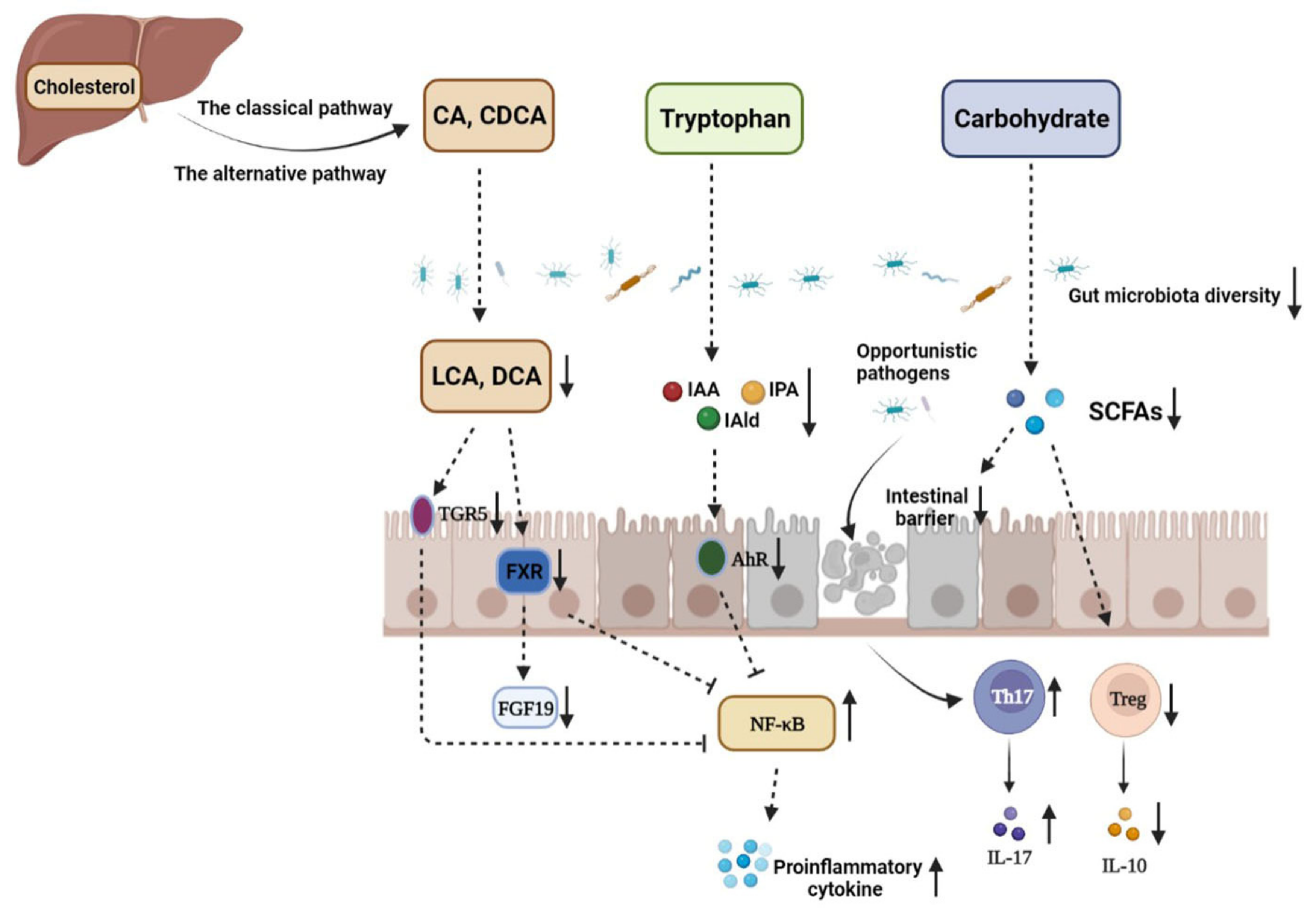
2. SCFAs and IBD
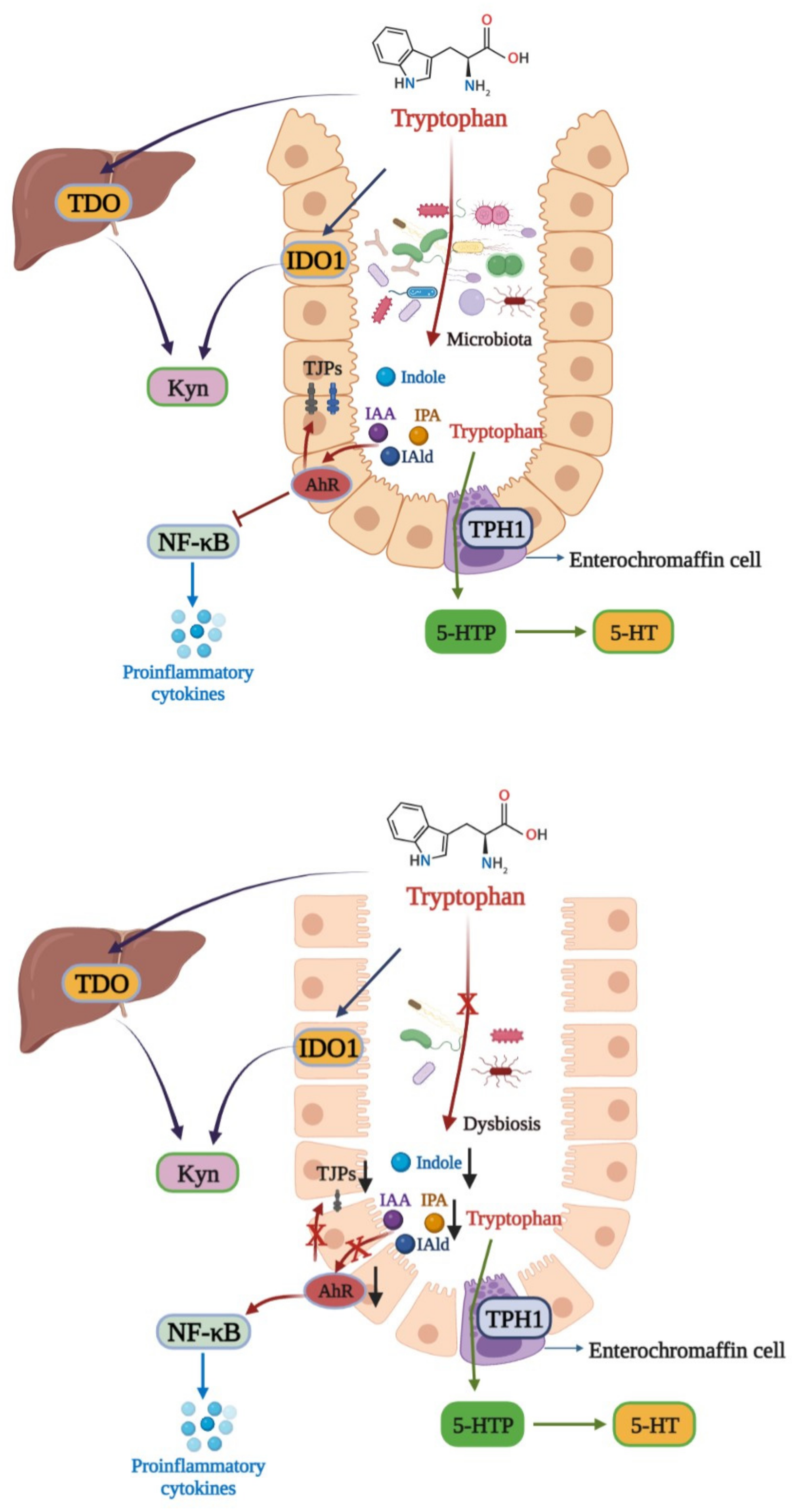
3. Tryptophan Metabolism and IBD
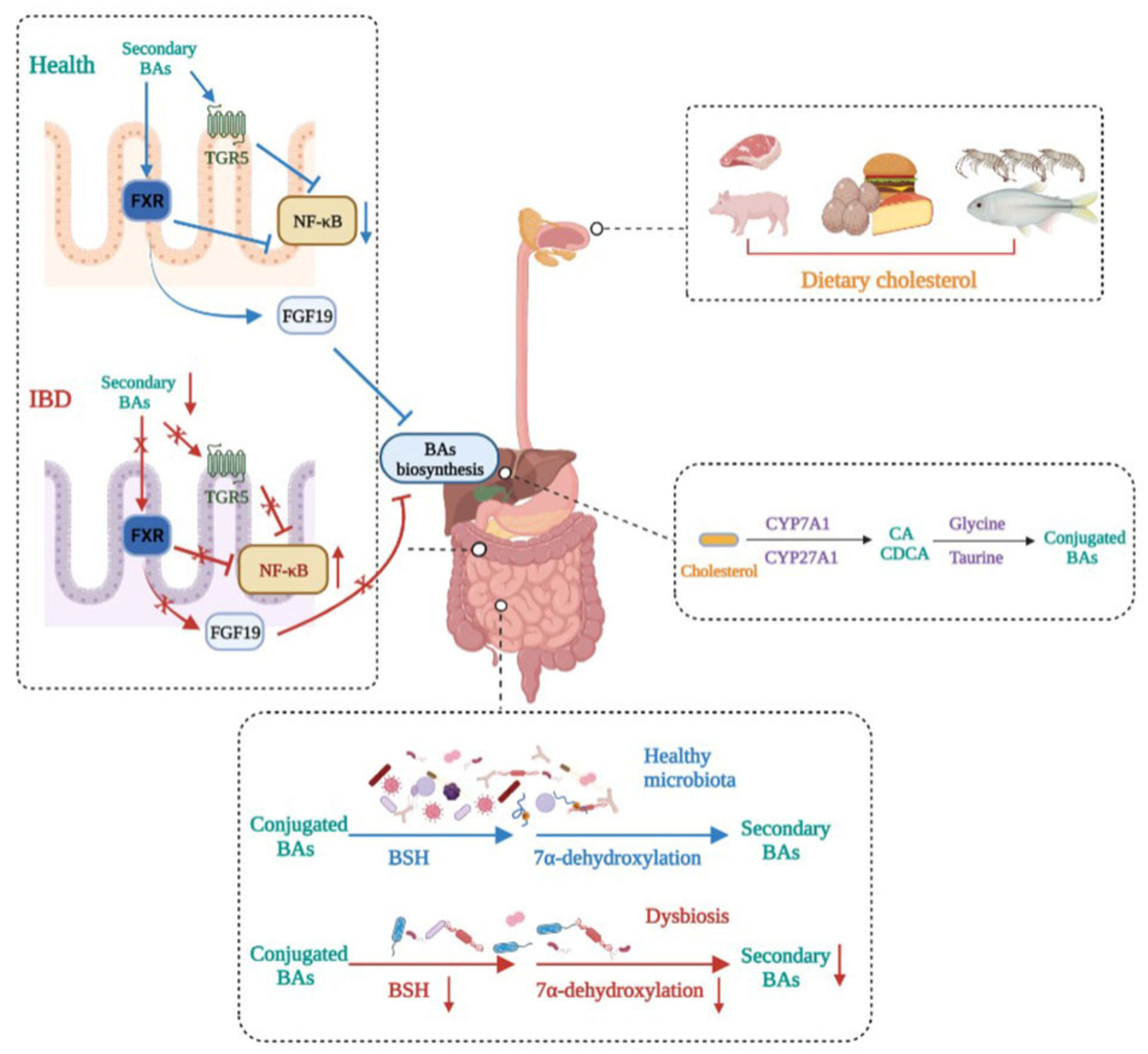
4. Bile Acids and IBD
This entry is adapted from the peer-reviewed paper 10.3390/nu14235140
References
- Atreya, R.; Siegmund, B. Location is important: Differentiation between ileal and colonic Crohn’s disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 544–558.
- Lophaven, S.N.; Lynge, E.; Burisch, J. The incidence of inflammatory bowel disease in Denmark 1980–2013: A nationwide cohort study. Aliment. Pharmacol. Ther. 2017, 45, 961–972.
- van den Heuvel, T.R.A.; Jeuring, S.F.G.; Zeegers, M.P.; van Dongen, D.H.E.; Wolters, A.; Masclee, A.A.M.; Hameeteman, W.H.; Romberg-Camps, M.J.L.; Oostenbrug, L.E.; Pierik, M.J.; et al. A 20-Year Temporal Change Analysis in Incidence, Presenting Phenotype and Mortality, in the Dutch IBDSL Cohort-Can Diagnostic Factors Explain the Increase in IBD Incidence? J. Crohns Colitis 2017, 11, 1169–1179.
- Mak, W.Y.; Zhao, M.; Ng, S.C.; Burisch, J. The epidemiology of inflammatory bowel disease: East meets west. J. Gastroenterol. Hepatol. 2020, 35, 380–389.
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584.
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247.
- Halme, L.; Paavola-Sakki, P.; Turunen, U.; Lappalainen, M.; Farkkila, M.; Kontula, K. Family and twin studies in inflammatory bowel disease. World J. Gastroenterol. 2006, 12, 3668–3672.
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217.
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608.
- Nakase, H.; Uchino, M.; Shinzaki, S.; Matsuura, M.; Matsuoka, K.; Kobayashi, T.; Saruta, M.; Hirai, F.; Hata, K.; Hiraoka, S.; et al. Evidence-based clinical practice guidelines for inflammatory bowel disease 2020. J. Gastroenterol. 2021, 56, 489–526.
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24.
- Lo Sasso, G.; Khachatryan, L.; Kondylis, A.; Battey, J.N.D.; Sierro, N.; Danilova, N.A.; Grigoryeva, T.V.; Markelova, M.I.; Khusnutdinova, D.R.; Laikov, A.V.; et al. Inflammatory Bowel Disease-Associated Changes in the Gut: Focus on Kazan Patients. Inflamm. Bowel Dis. 2021, 27, 418–433.
- Yu, M.; Wang, Q.M.; Ma, Y.H.; Li, L.Z.; Yu, K.; Zhang, Z.C.; Chen, G.Q.; Li, X.S.; Xiao, W.D.; Xu, P.Y.; et al. Aryl Hydrocarbon Receptor Activation Modulates Intestinal Epithelial Barrier Function by Maintaining Tight Junction Integrity. Int. J. Biol. Sci. 2018, 14, 69–77.
- Bedford, A.; Gong, J. Implications of butyrate and its derivatives for gut health and animal production. Anim. Nutr. 2018, 4, 151–159.
- Sasaki, K.; Inoue, J.; Sasaki, D.; Hoshi, N.; Shirai, T.; Fukuda, I.; Azuma, T.; Kondo, A.; Osawa, R. Construction of a Model Culture System of Human Colonic Microbiota to Detect Decreased Lachnospiraceae Abundance and Butyrogenesis in the Feces of Ulcerative Colitis Patients. Biotechnol. J. 2019, 14, 1800555.
- Zhou, L.X.; Zhang, M.M.; Wang, Y.M.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.T.; Tang, D.H.; Xu, L.; Yin, Y.Y.; et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940.
- Zhu, L.; Xu, L.-Z.; Zhao, S.; Shen, Z.-F.; Shen, H.; Zhan, L.-B. Protective effect of baicalin on the regulation of Treg/Th17 balance, gut microbiota and short-chain fatty acids in rats with ulcerative colitis. Appl. Microbiol. Biotechnol. 2020, 104, 5449–5460.
- Xu, H.M.; Zhao, H.L.; Guo, G.J.; Xu, J.; Zhou, Y.L.; Huang, H.L.; Nie, Y.Q. Characterization of short-chain fatty acids in patients with ulcerative colitis: A meta-analysis. BMC Gastroenterol. 2022, 22, 117.
- Kanda, N.; Hoashi, T.; Saeki, H. The Defect in Regulatory T Cells in Psoriasis and Therapeutic Approaches. J. Clin. Med. 2021, 10, 3880.
- Sun, M.M.; Wu, W.; Liu, Z.J.; Cong, Y.Z. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8.
- Gao, Y.A.; Davis, B.; Zhu, W.S.; Zheng, N.; Meng, D.; Walker, W.A. Short-chain fatty acid butyrate, a breast milk metabolite, enhances immature intestinal barrier function genes in response to inflammation in vitro and in vivo. Am. J. Physiol.-Gastrointest. Liver Physiol. 2021, 320, G521–G530.
- Zhang, X.; Tong, Y.; Lyu, X.; Wang, J.; Wang, Y.; Yang, R. Prevention and Alleviation of Dextran Sulfate Sodium Salt-Induced Inflammatory Bowel Disease in Mice With Bacillus subtilis-Fermented Milk via Inhibition of the Inflammatory Responses and Regulation of the Intestinal Flora. Front. Microbiol. 2020, 11, 622354.
- Peng, Y.; Yan, Y.; Wan, P.; Chen, D.; Ding, Y.; Ran, L.; Mi, J.; Lu, L.; Zhang, Z.; Li, X.; et al. Gut microbiota modulation and anti-inflammatory properties of anthocyanins from the fruits of Lycium ruthenicum Murray in dextran sodium sulfate-induced colitis in mice. Free Radic. Biol. Med. 2019, 136, 96–108.
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724.
- Rothhammer, V.; Quintana, F.J. The aryl hydrocarbon receptor: An environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol. 2019, 19, 184–197.
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.E.; Mayo, L.; Chao, C.C.; Patel, B.; Yan, R.; Blain, M.; et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586–597.
- Singh, R.; Chandrashekharappa, S.; Bodduluri, S.R.; Baby, B.V.; Hegde, B.; Kotla, N.G.; Hiwale, A.A.; Saiyed, T.; Patel, P.; Vijay-Kumar, M.; et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat. Commun. 2019, 10, 89.
- Jing, W.H.; Dong, S.J.; Luo, X.L.; Liu, J.J.; Wei, B.; Du, W.; Yang, L.; Luo, H.; Wang, Y.T.; Wang, S.C.; et al. Berberine improves colitis by triggering AhR activation by microbial tryptophan catabolites. Pharmacol. Res. 2021, 164, 105358.
- Ding, X.; Bin, P.; Wu, W.; Chang, Y.; Zhu, G. Tryptophan Metabolism, Regulatory T Cells, and Inflammatory Bowel Disease: A Mini Review. Mediat. Inflamm. 2020, 2020, 9706140.
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237.
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Hasler, R.; et al. Increased Tryptophan Metabolism Is Associated With Activity of Inflammatory Bowel Diseases. Gastroenterology 2017, 153, 1504–1516.e2.
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; MacDonald, T.T.; Pallone, F.; Monteleone, G. Aryl Hydrocarbon Receptor-Induced Signals Up-regulate IL-22 Production and Inhibit Inflammation in the Gastrointestinal Tract. Gastroenterology 2011, 141, U237–U324.
- Furumatsu, K.; Nishiumi, S.; Kawano, Y.; Ooi, M.; Yoshie, T.; Shiomi, Y.; Kutsumi, H.; Ashida, H.; Fujii-Kuriyama, Y.; Azuma, T.; et al. A Role of the Aryl Hydrocarbon Receptor in Attenuation of Colitis. Dig. Dis. Sci. 2011, 56, 2532–2544.
- Kim, H.Y.; Yoo, T.H.; Cho, J.Y.; Kim, H.C.; Lee, W.W. Indoxyl sulfate-induced TNF-alpha is regulated by crosstalk between the aryl hydrocarbon receptor, NF-kappa B, and SOCS2 in human macrophages. FASEB J. 2019, 33, 10844–10858.
- Scott, S.A.; Fu, J.J.; Chang, P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 19376–19387.
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294.
- Natividad, J.M.; Agus, A.; Planchais, J.; Lamas, B.; Jarry, A.C.; Martin, R.; Michel, M.-L.; Chong-Nguyen, C.; Roussel, R.; Straube, M.; et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018, 28, 737–749.e734.
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128.
- Iracheta-Vellve, A.; Calenda, C.D.; Petrasek, J.; Ambade, A.; Kodys, K.; Adorini, L.; Szabo, G. FXR and TGR5 Agonists Ameliorate Liver Injury, Steatosis, and Inflammation After Binge or Prolonged Alcohol Feeding in Mice. Hepatol. Commun. 2018, 2, 1379–1391.
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539.
- Agus, A.; Clement, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182.
- Komohara, Y.; Fujiwara, Y.; Ohnishi, K.; Takeya, M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv. Drug Deliv. Rev. 2016, 99, 180–185.
- Mańkowska-Wierzbicka, D.; Stelmach-Mardas, M.; Gabryel, M.; Tomczak, H.; Skrzypczak-Zielińska, M.; Zakerska-Banaszak, O.; Sowińska, A.; Mahadea, D.; Baturo, A.; Wolko, Ł.; et al. The Effectiveness of Multi-Session FMT Treatment in Active Ulcerative Colitis Patients: A Pilot Study. Biomedicines 2020, 8, 268.
