Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The constant rise in the global temperature and unpredictable shifts in precipitation patterns are two of the main effects of climate change. Conifers originated more than 300 million years ago and currently dominate many temperate and boreal forests.
- gymnosperms
- molecular
- next-generation breeding
- water stress
- genetics
- genomic
- drought resistance
- Picea
- Abies
- Pinus
1. Introduction
According to the FAO’s Global Forest Resources Assessment 2020 (https://www.fao.org/documents/card/en/c/ca8753en (accessed on 2 July 2022)), 4.06 billion hectares of land are covered by forests worldwide, corresponding to over 30% of the total land area. Over the last decades, climate change has increased the risk of drought stress (DS) in many regions, mainly via increased temperatures, altered precipitation patterns and faster snow melt [1][2]. The effects of droughts on forest plants vary, from reduced growth in cases of moderate droughts to mass mortality if severe droughts occur [3][4]. Global simulations predict widespread massive tree mortality under the projected rise in global temperatures and extremes that accompanies drought [5]. Therefore, it is of primary importance to understand plant response and adaptation mechanisms to drought in order to properly manage tree populations and select those individuals or provenances that show a higher resistance level. Drought resistance is a very complex trait with both environmental and genetic components, and different populations of the same species may not respond equally to a given climate [6][7]. Moreover, several distinct aspects can be important when drought tolerance (DT) is concerned. As an example, in regards to wood production, the capability of the plant to grow in low-water conditions might be the most important trait to consider, while for a natural forest, adaptations to highly variable water availability might be more favorable. In all cases, the main target of research studies is to understand what traits are more important to achieving drought resistance and to find the relationship between phenotypes and genotypes [8][9].
Conifers originated more than 300 million years ago and currently dominate many temperate and boreal forests. They have very large genomes (18 to 35 Gb), showing a very different structure and composition when compared with those of angiosperm genomes. Despite an apparently conserved genome structure, conifers demonstrate some competitive capacity, as different taxa are adapted to a wide variety of environmental conditions [10]. Even though angiosperms and gymnosperms share some general principles regarding drought resistance, there are also significant differences. Some studies suggest that angiosperm tree species, in general, tend to be less sensitive to drought than gymnosperm species [11][12]. At first glance, one of the fundamental differences in water transport is the size and function of conifer tracheids and angiosperm vessels [13]. In fact, even though conifers have greater stem hydraulic safety than angiosperms, during drought events, conifers experience more frequent embolisms than angiosperms in distal tissues [14]. Embolism repair is likely driven by sugars that come from nearby parenchyma cells, and conifers have few carbohydrates or parenchyma in their xylems compared to angiosperms. So, even though conifers tend to experience embolism more frequently in their leaves and roots than angiosperms, these organs may act as hydraulic circuit breakers to prevent stem embolism in conifers [15].
2. Plant Strategies to Cope with Drought
Intuitively, a very simple definition of DT is the ability of a plant to survive a prolonged period of water shortage. This can be true in particular environments, such as deserts or semi-arid regions where water availability is low most of the time. For the analysis of DT in more temperate areas, different climatic drivers and levels of variability among tree species should be taken into account [16]. Among other factors, trees’ growth and in particular trees’ short-term responses to extreme drought events are two of the most frequently studied [17]. Plants that are adapted to low water levels can also show a very low growth rate in favorable conditions as a consequence of more conservative resource usage, and therefore they may not be competitive with other, less conservative species [18][19]. Moreover, some studies on Abies concolor, Pinus lambertiana and P. sylvestris L. have correlated slow growth and sudden decreases in growth with a higher mortality [20][21].
Therefore, growth plasticity, the capability of plants to display a high growth rate in good climatic conditions and a low growth rate in drought conditions, can be considered a very favorable trait, especially in variable environments [22].
Different types of DS actually exist. A single extreme episode of drought is a different type of stress compared to the stress resulting from low but constant water availability. Moreover, a different kind of stress results from multiple drought episodes occurring during a growing season, and even a single drought event can occur in different periods of a plant’s growing season. Therefore, plant response is very complex, and multiple strategies should be adopted to cope with drought. Such strategies can be divided into three main groups: drought avoidance, drought resistance and drought resilience (Figure 1).
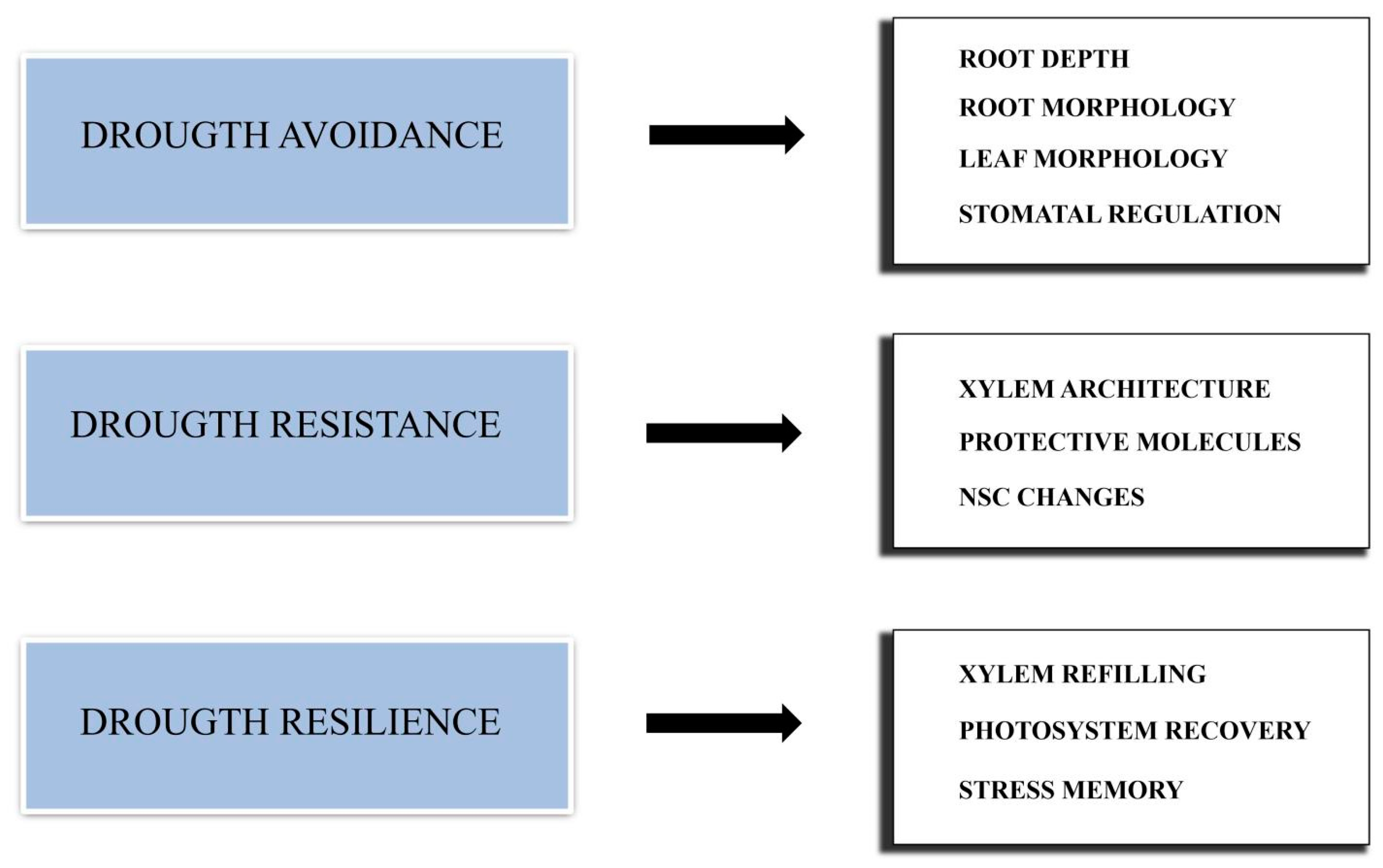
Figure 1. Schematic representation of the main strategies adopted by conifers to cope with drought stress.
3. Studying Drought Tolerance in Conifers
In a context of climate change, in which drought events are predicted to increase in frequency and severity in the near future [23], it is necessary to understand how plants are capable of adapting to low water availability. This is particularly true for conifers, which are often a dominant component of arid zone forests. Plant adaptations to drought can be studied at different levels. The main approaches consider traits at anatomical/morphological, physiological/biochemical and genetic levels.
3.1. Anatomical and Morphological Studies
Morphological studies usually link a specific phenotype to DT. In conifers, tree growth, often measured by the basal area increment or tree ring width, is a widely used parameter that can be correlated to meteorological data to assess plants’ responses to drought events [24]. Tree growth measurements have been applied successfully to understand the effects of forest management on the drought responses of several species, such as Pinus nigra [25], Pinus halepensis [26] and Pinus sylvestris [27][28]. The basal area increment was coupled with shoot elongation measures to compare the drought responses of adult trees and saplings in three different Pinus species, in order to predict future scenarios of relic forests under climate change [29]. In another study, the stem radial growth was used to compare the responses of Pinus edulis (isohydric species) and Juniperus monosperma (anisohydric species) to DS [30]. Tree height can be used as a morphological characteristic to study conifers’ response to drought. In a natural forest, the relationships between dominant trees and smaller ones may vary with climate change. When an ample supply of water is guaranteed, light can be the main limiting factor for plant growth, so the dominant trees are favored. In contrast, when the water availability becomes a limiting factor, the more shaded positions of smaller trees may limit transpiration and therefore compensate for the lack of light [31].
Another important morphological characteristic that is associated with drought resistance is the xylem morphology, in particular the lumen diameter and the thickness of the tracheid walls. These characteristics largely influence the xylem’s water transport efficiency as well as its safety, which influence the probability of tracheid implosions during water stress [32]. Overall, conifers are capable of modifying the xylem’s structure in response to droughts [33]. An arid climate seems to promote xylem efficiency over safety [34][35]. In a recent study on Picea abies, a macroscopic characteristic, namely stem cracks, was associated with water stress [36]. In the same study, the thickness of the tracheid walls, rather than the lumen diameter, was found to be the main anatomical characteristic associated with tracheid collapses. This finding is particularly important for all tree species exploited for wood production, as the cell wall thickness was shown to have quite a high heritability [37]. Cell wall thickness is a parameter determining wood density, and wood density has been negatively correlated to growth in Picea abies [38]. The xylem phenology was also studied in conifers in relationship to drought [39]. Characteristics, such as cell differentiation, cell enlargement and cell wall thickening, seem to be influenced by water availability in several species, such as Abies alba, Pinus sylvestris [40] and Larix decidua [41].
When studying DT in plants, one should take into account that natural populations are not homogeneous but present a certain degree of genetic variation. Morphological traits, such as the basal area increment or tree ring width, can be used to assess the inter- and intraspecific genetic variation. This is usually performed in so-called provenance or common garden studies, in which seedlings from many different regions can be planted and studied in a common environment [42][43]. Provenance studies can be conducted across multiple sites or using multiple treatments in order to estimate the plasticity of traits [44]. The intraspecific growth response to drought was studied in Abies alba using provenances from Bulgaria, Italy, Romania and Czech Republic. The ring width, earlywood width and latewood width were measured and correlated to drought events over a period of over 20 years, in order to find provenances combining a high productivity and drought resistance [45]. In a second study, 43 populations of Picea glauca were evaluated for DT by measuring a series of tree-ring traits. A significant genetic variation was found among populations in response to DS. In particular, the authors found that populations from drier geographical origins showed a higher resilience to extreme drought events when compared to populations from more humid geographical origins, indicating local genetic adaptations [46]. Similar experiments were performed using several phenotypical traits on different conifer species, such as Pinus pinea [47], Pinus ponderosa [48], Pinus sylvestris [49] and Picea abies [50]. An interspecific study was performed comparing the drought responses of Picea abies, Abies alba, Larix decidua and Pseudotsuga menziesii. This latter species had the highest drought resistance, while Abies alba had the best drought recovery. Nonetheless, even the most drought-sensitive species, Picea abies and Larix decidua, showed significant genetic variation within and among populations along their natural geographic areas, enough to justify targeted tree breeding and supportive forest management [51].
3.2. Physiological and Biochemical Studies
Most of the studies using morphological data to assess DT are focused on tree growth. This parameter is a good indicator of plant resilience, and it is very useful when wood production is the final goal. Nevertheless, DT is a very complex trait involving a number of different mechanisms. In order to identify such mechanisms and use them to highlight differences in plant responses to drought, physiological datacan be used [52][53]. One of the most studied physiological processes related to water stress is photosynthesis [54][55][56]. In particular, the concentration of chlorophyll a can be obtained via the extrapolation of the emission of refracted light from foliage [57], therefore it is a parameter that can be measured by remote-sensing tools and can be used to monitor large areas [58][59]. Drought can damage Photosystem II (PSII), resulting in changes in fluorescence parameters [60]. The fluorescence measurement was shown to be a very sensitive proxy for DS [61], allowing researchers to assess physiological disturbances even before the appearance of visible symptoms [60]. In the last few years, several studies on conifers have been published [62][63]. In Picea abies, the chlorophyll a concentration and fluorescence parameters were measured together with the tree height to assess seedling performances under water stress. Both physiological parameters were good indicators of plants’ drought sensitivity, even though differences were found depending on the soil type [64]. In Abies alba, the chlorophyll a fluorescence was measured, testing five provenances from different altitudes under mild water stress. Significant differences were found, with provenances from higher altitudes showing better performances under both optimal and low-water conditions, suggesting that there were local adaptations to drought and that fluorescence parameters can be applied during plant selection for resistant seedlings [65].
Another important physiological parameter that has been often used to assess plant DT is water use efficiency (WUE), which is the ratio between the carbon fixed by photosynthesis and the water loss [66][67][68]. Nevertheless, caution must be taken when using WUE as an indicator for DT, because it depends on different mechanisms, such as photosynthesis and transpiration, which can both vary and not always in the same way.
Moreover, several measures of WUE exist and even if they are often correlated, they are not interchangeable [69], so they may lead to contrasting results. One of the most frequently used methods for measuring WUE is the carbon ratio δ13 C [70]. In conifers, some studies have found higher δ13 C in populations originating from drier sites [18], while others have shown the opposite [71]. In Pinus halepensis, individuals from dry sites showed a lower WUE plasticity than those from mesic sites [67], while in another study, a higher average WUE was shown in individuals from drier sources [66]. Finally, it must be noted that, especially if measured during the whole growing season, high water usage due to highly plastic growth can reduce the WUE, even though plastic growth can be considered a desirable characteristic in moderately dry climates [18]. For all these reasons, it is always advisable to integrate WUE with other parameters when dealing with drought resistance [72][73].
3.3. Molecular Genetics Studies
To further investigate drought responses and link morphological and physiological traits to the genes and/or genomic regions that are responsible for these characteristics, molecular genetics can be used [74]. One of the most applied approaches is a gene expression analysis. Although there are a lot of techniques to study gene expression, in the most recent publications, whole-transcriptome approaches [75][76][77][78][79][80] and/or quantitative real-time PCR (qRT-PCR) [77][78][81] have been used more frequently. In contrast to RNAseq and other whole-transcriptome techniques, qRT-PCR is very sensitive but in most cases it is used to study one or a few specific target genes [82] or to confirm the gene expression results obtained with less sensitive methods [78][83]. Via expression analyses, a range of genes have been identified that might be involved in the drought responses in conifers.
Genes related to signaling and transcription factors, including AP2/ERF, bZIP, TCP, WRKY and MYB, have been found to be regulated during water stress in several conifer species, such as Larix kaempferi [84], Pinus massoniana [85], Abies alba [83] and Pinus taeda [86]. All these genes, due to their regulatory function, are usually expressed quite early during DS and therefore they could be considered good indicators of an efficient plant response. Nevertheless, this is not always the case. In Pinus pinaster, it was found that tolerant individuals can be pre-adapted to cope with drought, constitutively expressing stress-related genes; in contrast, in more sensitive individuals, these are induced by the onset of stress [76]. So, care must be taken when considering gene expression, and distinguishing between the different drivers of observed differences is of primary importance. Another important factor that must be considered when studying gene expression during drought is the type of treatment used. In some studies, DS was induced by stopping irrigation [79]; in others, a chemically-induced stress was used [87]. In some cases, the water was withheld for a given period [87]; in others, it was withheld until the needles wilted [86]. Thus, there is always the possibility of methodological artifacts [88]. Abscisic acid (ABA), a plant hormone involved in stomatal closure, shoot growth and water uptake, was shown to regulate many structural genes in conifers [78][79][85] even though ABA-independent pathways also exist in many species [89]. Other genes regulated during drought include those encoding for antioxidants [75][84], protective molecules, such as late embyogenesis abundant (LEA) proteins, which are thought to stabilize proteins and membranes [90], genes involved in lignin and sugar biosynthesis [84], flavonoid and terpenoid biosynthesis [75], aquaporins, which can affect the water permeability of membranes [85] and even pathogen resistance genes, such as nucleotide-binding, leucine-rich repeat proteins [91][92].
4. Next Generation Breeding
The increasing amount of morphological, physiological and molecular data on conifer drought resistance can have practical applications in improving breeding programs [93][94][95][96] (Figure 2).
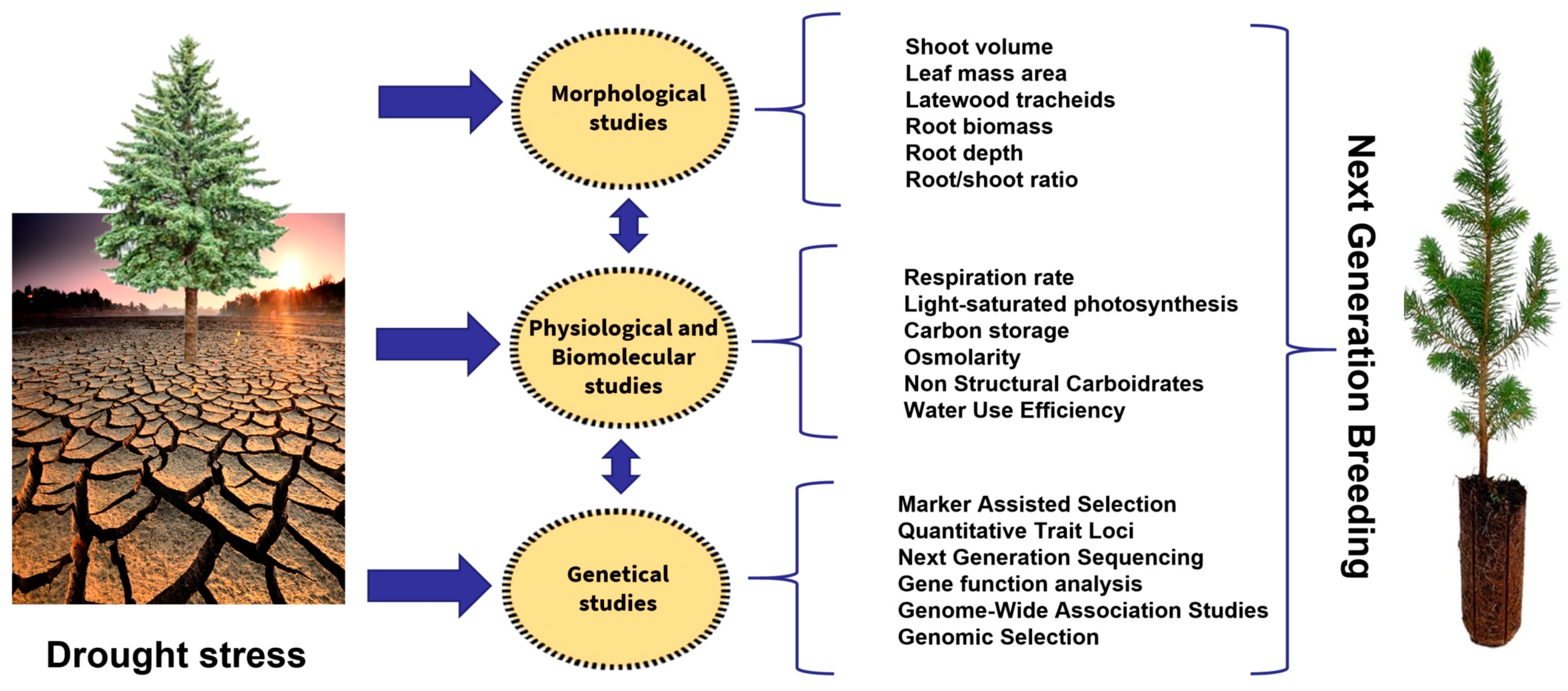
Figure 2. Next-generation breeding processes to improve drought stress in conifers.
As a matter of fact, traditional breeding programs for long-living trees, such as conifers, face a number of challenges. The most important is the great amount of space and time necessary to obtain significant achievements. Even though several breeding programs started in the 1950s, many of them are only in their first or second cycle of selection, with the most advanced ones, such as the loblolly pine (Pinus taeda L.) breeding program in the USA, in the third or fourth cycles [97]. Moreover, due to the high costs, tree breeding programs have been developed mainly for commercially important species and only recently has it become evident that it is important to also protect natural forests, which are often threatened by climate change [98]. In cases of drought, plant resistance or tolerance is controlled by multiple genes, each with a minor additive effect, so it is particularly challenging to achieve and maintain the desired level of resistance [99].
Another important factor that must be taken into account, especially when considering natural forests, is genetic diversity. Resistant populations developed by breeding programs have to maintain all the adaptive traits typical of the species or provenance, such as growth rate, cold resistance and pathogen resistance. Therefore, it is necessary to exclude the presence of negative correlations between drought resistance and other adaptive traits [97].
New emerging technologies can be efficiently used to link plant genome to phenotypic data and therefore could be used to improve the efficiency of traditional breeding and accelerate the selection of valuable genotypes. Nowadays, high-throughput technologies can be applied not only to genotyping but also to phenotyping, improving both data acquisition and analytical pipelines, as well as possibly leading to an unprecedented revolution in the way has been studied agriculture till now [100]. As an example, a number of portable and user-friendly chlorophyll fluorimeters have been introduced in the last few years. These, together with more sophisticated high-throughput hyperspectral imaging systems, have enhanced the accuracy of chlorophyll measurements. In this way, large phenotyping platforms have been implemented with an automatic control and data analysis system, allowing researchers to keep a large number of plants in parallel under constant monitoring, over long periods of time and in different environmental conditions [101]. These high-throughput phenotyping technologies are generally employed in controlled environments, such as growth chambers and greenhouses, so there are some limitations when predicting the performance of a plant under field conditions. Nonetheless, these systems can be very useful in studying how phenotypes change among different genotypes under uniform stress conditions [102].
A large amount of genomic resources is available for conifers. Despite their size and complexity, several conifer genomes have been sequenced [103][104][105][106][107][108]. A complete list of genomic resources of conifers, with the year of release, the Genebank storage and the accession number, has been reported by Traversari et al. [109]. These data, together with transcriptome studies, have provided useful information for the development of high-density genetic maps [110][111][112][113][114] and SNP arrays [115][116][117][118][119], which could be used in next-generation breeding approaches. To date, marker-assisted selection (MAS) has shown only a limited application in forest breeding [120][121]. The main reason is that MAS is quite reliable and easy to use in cases of monogenic characteristics, but it is much harder to be applied in cases of more complex traits, such as drought, as several markers linked to genetic loci involved in the control of the resistance must be identified. As already stated (see previous paragraph), classic QTL mapping had little success in conifers for the identification of genetic loci linked to traits of interest. Moreover, each locus can control a relatively small percentage of the resistance, so several loci should be screened at the same time in order to reach the desired level of resistance and should be validated using different populations before being routinely used for assisted selection [43][122]. Overall, it was shown that QTLs are not effectively suitable for MAS in forest trees, as they do not explain enough genetic variations for complex traits, such as drought resistance [123].
This entry is adapted from the peer-reviewed paper 10.3390/f13122016
References
- Bussotti, F.; Pollastrini, M. Revisiting the concept of stress in forest trees at the time of global change and issues for stress moni-toring. Plant Stress 2021, 2, 100013.
- Caballero, C.B.; Ruhoff, A.; Biggs, T. Land use and land cover changes and their impacts on surface-atmosphere interactions in Brazil: A systematic review. Sci. Total. Environ. 2022, 808, 152134.
- Lloret, F.; Jaime, L.A.; Margalef-Marrase, J.; Perez-Navarro, M.A.; Batllori, E. Short-term forest resilience after drought-induced die-off in Southwestern European forests. Sci. Total Environ. 2022, 806, 150940.
- Navarro-Cerrillo, R.M.; Gonzalez-Moreno, P.; Ruiz-Gomez, F.J.; Sanchez-Cuesta, R.; Gazol, A.; Camarero, J.J. Drought stress and pests increase defoliation and mortality rates in vulnerable Abies pinsapo forests. For. Ecol. Manag. 2022, 504, 119824.
- McDowell, N.G.; Williams, A.P.; Xu, C.; Pockman, W.T.; Dickman, L.T.; Sevanto, S.; Pangle, R.; Limousin, J.; Plaut, J.; Mackay, D.S.; et al. Multi-scale predictions of massive conifer mortality due to chronic temperature rise. Nat. Clim. Change 2016, 6, 295–300.
- Rehfeldt, G.E.; Leites, L.P.; St Clair, J.B.; Jaquish, B.C.; Saenz-Romero, C.; Lopez-Upton, J.; Joyce, D.G. Comparative genetic re-sponses to climate in the varieties of Pinus ponderosa and Pseudotsuga menziesii: Clines in growth potential. For. Ecol. Manag. 2014, 324, 138–146.
- Rehfeldt, G.E.; Tchebakova, N.M.; Parfenova, Y.I.; Wykoff, W.R.; Kuzmina, N.A.; Milyutin, L.I. Intraspecific responses to climate in Pinus sylvestris. Glob. Change Biol. 2002, 8, 912–929.
- Reinhardt, K.; Germino, M.; Kueppers, L.M.; Domec, J.-C.; Mitton, J. Linking carbon and water relations to drought-induced mortality in Pinus flexilis seedlings. Tree Physiol. 2015, 35, 771–782.
- Sevanto, S.; McDowell, N.G.; Dickman, L.T.; Pangle, R.; Pockman, W.T. How do trees die? A test of the hydraulic failure and car-bon starvation hypotheses. Plant Cell Environ. 2014, 37, 153–161.
- Diaz-Sala, C.; Cabezas, J.A.; Fernández de Simón, B.; Abarca, D.; Guevara, M.Á.; de Miguel, M.; Cadahía, E.; Aranda, I.; Cervera, M.T. The uniqueness of conifers. In From Plant Genomics to Plant Biotechnology, 1st ed.; Poltronieri, P., Burbulis, N., Fogher, C., Eds.; Woodhead Publishing: Sawston, UK, 2013; pp. 67–96.
- Mitchell, P.J.; O’Grady, A.P.; Tissue, D.T.; Worledge, D.; Pinkard, E.A. Co-ordination of growth, gas exchange and hydraulics define the carbon safety margin in tree species with contrasting drought strategies. Tree Physiol. 2014, 34, 443–458.
- Anderegg, W.R.; Schwalm, C.; Biondi, F.; Camarero, J.J.; Koch, G.; Litvak, M.; Wolf, A. Pervasive drought legacies in forest eco-systems and their implications for carbon cycle models. Science 2015, 349, 528–532.
- Sperry, J.S.; Hacke, U.G.; Pittermann, J. Size and function in conifer tracheids and angiosperm vessels. Am. J. Bot. 2006, 93, 1490–1500.
- Rosner, S.; Johnson, D.M.; Voggeneder, K.; Domec, J.C. The conifer-curve: Fast prediction of hydraulic conductivity loss and vulnerability to cavitation. Ann. For. Sci. 2019, 76, 82.
- Johnson, D.M.; McCulloh, K.A.; Woodruff, D.R.; Meinzer, F.C. Hydraulic safety margins and embolism reversal in stems and leaves: Why are conifers and angiosperms so different? Plant Sci. 2012, 195, 48–53.
- Zang, C.; Hartl-Meier, C.; Dittmar, C.; Rothe, A.; Menzel, A. Patterns of drought tolerance in major European temperate forest trees: Climatic drivers and levels of variability. Glob. Change Biol. 2014, 20, 3767–3779.
- Eilmann, B.; Rigling, A. Tree-growth analyses to estimate tree species’ drought tolerance. Tree Physiol. 2012, 32, 178–187.
- Kerr, K.L.; Meinzer, F.C.; McCulloh, K.A.; Woodruff, D.R.; Marias, D.E. Expression of functional traits during seedling establish-ment in two populations of Pinus ponderosa from contrasting climates. Tree Physiol. 2015, 35, 535–548.
- Montwé, D.; Spiecker, H.; Hamann, A. Five decades of growth in a genetic field trial of Douglas-fir reveal trade-offs between productivity and drought tolerance. Tree Genet. Genomes 2015, 11, 29.
- Das, A.J.; Battles, J.J.; Stephenson, N.L.; van Mantgem, P.J. The relationship between tree growth patterns and likelihood of mortality: A study of two tree species in the Sierra Nevada. Can. J. For. Res. 2007, 37, 580–597.
- Hereş, A.M.; Martínez-Vilalta, J.; Claramunt López, B. Growth patterns in relation to drought-induced mortality at two Scots pine (Pinus sylvestris L.) sites in NE Iberian Peninsula. Trees 2012, 26, 621–630.
- Santos-Del-Blanco, L.; Bonser, S.P.; Valladares, F.; Chambel, M.R.; Climent, J. Plasticity in reproduction and growth among 52 range-wide populations of a Mediterranean conifer: Adaptive responses to environmental stress. J. Evol. Biol. 2013, 26, 1912–1924.
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Hogg, E.H.; et al. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. Forest Ecol. Manag. 2010, 259, 660–684.
- Proutsos, N.; Tigkas, D. Growth response of endemic black pine trees to meteorological variations and drought episodes in a mediterranean region. Atmosphere 2020, 11, 554.
- Lucas-Borja, M.E.; Andivia, E.; Candel-Pérez, D.; Linares, J.; Camarero, J. Long term forest management drives drought resilience in Mediterranean black pine forest. Trees 2021, 35, 1651–1662.
- Manrique-Alba, À.; Beguería, S.; Molina, A.J.; González-Sanchis, M.; Tomàs-Burguera, M.; del Campo, A.D.; Colangelo, M.; Camarero, J.J. Long-term thinning effects on tree growth, drought response and water use efficiency at two Aleppo pine plantations in Spain. Sci. Total Environ. 2020, 728, 138536.
- Duque Lazo, J.; Navarro-Cerrillo, R.; Sanchez-Salguero, R.; Rodriguez Vallejo, C. Is thinning an alternative when trees could die in response to drought? The case of planted Pinus nigra and P. Sylvestris stands in southern Spain. Forest Ecol. Manag. 2019, 433, 313–324.
- Sun, S.; Lei, S.; Jia, H.; Li, C.; Zhang, J.; Meng, P. Tree-ring analysis reveals density-dependent vulnerability to drought in planted mongolian pines. Forests 2020, 11, 98.
- Andivia, E.; Ruiz-Benito, P.; Díaz-Martínez, P.; Carro-Martínez, N.; Zavala, M.A.; Madrigal-González, J. Inter-specific tolerance to recurrent droughts of pine species revealed in saplings rather than adult trees. Forest Ecol. Manag. 2020, 459, 117848.
- Manrique-Alba, À.; Sevanto, S.; Adams, H.D.; Collins, A.D.; Dickman, L.T.; Chirino, E.; Bellot, J.; McDowell, N.G. Stem radial growth and water storage responses to heat and drought vary between conifers with differing hydraulic strategies. Plant Cell Environ. 2018, 41, 1926–1934.
- Pretzsch, H.; Schütze, G.; Biber, P. Drought can favour the growth of small in relation to tall trees in mature stands of Norway spruce and European beech. For. Ecosyst. 2018, 5, 20.
- Venturas, M.D.; Sperry, J.S.; Hacke, U.G. Plant xylem hydraulics: What we understand, current research, and future challenges. J. Integr. Plant Biol. 2017, 59, 356–389.
- Castagneri, D.; Fonti, P.; von Arx, G.; Carrer, M. How does climate influence xylem morphogenesis over the growing season? Insights from long-term intra-ring anatomy in Picea abies. Ann. Bot. 2017, 119, 1011–1020.
- Guérin, M.; von Arx, G.; Martin-Benito, D.; Andreu-Hayles, L.; Griffin, K.L.; McDowell, N.G.; Pockman, W.; Gentine, P. Distinct xylem responses to acute vs prolonged drought in pine trees. Tree Physiol. 2020, 40, 605–620.
- Martin-Benito, D.; Anchukaitis, K.J.; Evans, M.N.; Del Río, M.; Beeckman, H.; Cañellas, I. Effects of drought on xylem anatomy and water-use efficiency of two co-occurring pine species. Forests 2017, 8, 332.
- Rosner, S.; Gierlinger, N.; Klepsch, M.; Karlsson, B.; Evans, R.; Lundqvist, S.-O.; Světlík, J.; Børja, I.; Dalsgaard, L.; Andreassen, K.; et al. Hydraulic and mechanical dysfunction of Norway spruce sapwood due to extreme summer drought in Scandinavia. Forest Ecol. Manag. 2018, 409, 527–540.
- Chen, Z.; Karlsson, B.; Mörling, T.; Olsson, L.; Mellerowicz, E.; Wu, H.; Lundqvist, S.-O.; Garcia Gil, R. Genetic analysis of fiber dimensions and their correlation with stem diameter and solid-wood properties in Norway spruce. Tree Genet. Genomes 2016, 12, 123.
- Chen, Z.; Garcia Gil, R.; Karlsson, B.; Lundqvist, S.-O.; Olsson, L.; Wu, H. Inheritance of growth and solid wood quality traits in a large Norway spruce population tested at two locations in southern Sweden. Tree Genet. Genomes 2014, 10, 1291–1303.
- Fajstavr, M.; Bednářová, E.; Nezval, O.; Giagli, K.; Gryc, V.; Vavrčík, H.; Horacek, P.; Urban, J. How needle phenology indicates the changes of xylem cell formation during drought stress in Pinus sylvestris L. Dendrochronologia 2019, 56, 125600.
- Larysch, E.; Stangler, D.F.; Nazari, M.; Seifert, T.; Kahle, H.P. Xylem phenology and growth response of European beech, Silver fir and Scots pine along an elevational gradient during the extreme drought year 2018. Forests 2021, 12, 75.
- Saderi, M.; Rathgeber, C.; Rozenberg, P.; Fournier, M. Phenology of wood formation in larch (Larix decidua Mill.) trees growing along a 1000-m elevation gradient in the French Southern Alps. Ann. For. Sci. 2019, 76, 89.
- Leites, L.P.; Rehfeldt, G.E.; Robinson, A.P.; Crookston, N.L.; Jaquish, B. Possibilities and limitations of using historic provenance tests to infer forest species growth responses to Climate Change. Nat. Resour. Model. 2012, 25, 409–433.
- Capblancq, T.; Lachmuth, S.; Fitzpatrick, M.C.; Keller, S.R. From common gardens to candidate genes: Exploring local adaptation to climate in red spruce. New Phytol. 2022.
- Rehfeldt, G.E.; Jaquish, B.; Saenz-Romero, C.; Joyce, D.; Leites, L.; Clair, J.; Upton, J. Comparative genetic responses to climate in the varieties of Pinus ponderosa and Pseudotsuga menziesii: Reforestation. Forest Ecol. Manag. 2014, 324, 147–157.
- Mihai, G.; Alexandru, A.; Stoica, E.; Birsan, M.V. Intraspecific growth response to drought of Abies alba in the southeastern Car-pathians. Forests 2021, 12, 387.
- Depardieu, C.; Girardin, M.P.; Nadeau, S.; Lenz, P.; Bousquet, J.; Isabel, N. Adaptive genetic variation to drought in a widely distributed conifer suggests a potential for increasing forest resilience in a drying climate. New Phytol. 2020, 227, 427–439.
- Pardos, M.; Calama, R. Responses of Pinus pinea seedlings to moderate drought and shade: Is the provenance a differential factor? Photosynthetica 2017, 56, 786–798.
- Warwell, M.V.; Shaw, R.G. Phenotypic selection on ponderosa pine seed and seedling traits in the field under three experimen-tally manipulated drought treatments. Evol. Appl. 2019, 12, 159–174.
- Vizcaíno-Palomar, N.; González-Muñoz, N.; González-Martínez, S.; Alia, R.; Garzón, M. Most southern Scots pine populations are locally adapted to drought for tree height growth. Forests 2019, 10, 555.
- Budeanu, M.; Apostol, E.; Besliu, E.; Crișan, V.; Petritan, A.M. Phenotypic Variability and differences in the drought response of Norway spruce pendula and pyramidalis half-sib families. Forests 2021, 12, 947.
- Schueler, S.; George, J.-P.; Karanitsch-Ackerl, S.; Mayer, K.; Klumpp, R.T.; Grabner, M. Evolvability of drought response in four native and non-native conifers: Opportunities for forest and genetic resource management in Europe. Front. Plant Sci. 2021, 12, 1304.
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol. Life Sci. 2015, 72, 673–689.
- Ghazghazi, H.; Riahi, L.; Yangui, I.; Messaoud, C.; Rzigui, T.; Nasr, Z. Effect of drought stress on physio-biochemical traits and secondary metabolites production in the woody species Pinus halepensis Mill. at a juvenile development stage. J. Sustain. Forest 2022, 41, 878–894.
- Chaves, M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2008, 103, 551–560.
- Rao, D.E.; Chaitanya, V. Photosynthesis and antioxidative defense mechanisms in deciphering drought stress tolerance of crop plants. Biol. Plant. 2016, 60, 201–218.
- Drake, J.E.; Power, S.A.; Duursma, R.A.; Medlyn, B.E.; Aspinwall, M.J.; Choat, B.; Creek, D.; Eamusc, D.; Maier, C.; Pfautsch, S.; et al. Stomatal and non-stomatal limitations of photosynthesis for four tree species under drought: A comparison of model formulations. Agric. For. Meteorol. 2017, 247, 454–466.
- Malenovský, Z.; Homolova, L.; Zurita-Milla, R.; Lukeš, P.; Kaplan, V.; Hanus, J.; Gastellu-Etchegorry, J.P.; Schaepman, M. Re-trieval of spruce leaf chlorophyll content from airborne image data using continuum removal and radiative transfer. Remote Sens. Environ. 2013, 131, 85–102.
- Chen, J.H.; Wang, S.Q.; Chen, B.; Li, Y.; Amir, M.; Ma, L.; Zhu, K.; Yang, F.T.; Wang, X.B.; Liu, Y.Y.; et al. Comparative analysis on the estimation of diurnal solar-induced chlorophyll fluorescence dynamics for a subtropical evergreen coniferous forest. Remote Sens. 2021, 13, 3143.
- Lausch, A.; Borg, E.; Bumberger, J.; Dietrich, P.; Heurich, M.; Huth, A.; Jung, A.; Klenke, R.; Knapp, S.; Mollenhauer, H.; et al. Un-derstanding forest health with remote sensing, Part III: Requirements for a scalable multi-source forest health monitoring net-work based on data science approaches. Remote Sens. 2018, 10, 1120.
- Kalaji, H.M.; Jajoo, A.; Oukarroum, A.; Brestic, M.; Zivcak, M.; Samborska, I.A.; Cetner, M.D.; Łukasik, I.; Goltsev, V.; Ladle, R.J. Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant. 2016, 38, 102.
- Bigras, F. Photosynthetic response of white spruce families to drought stress. New For. 2005, 29, 135–148.
- Maseyk, K.; Lin, T.; Cochavi, A.; Schwartz, A.; Yakir, D. Quantification of leaf-scale light energy allocation and photoprotection processes in a Mediterranean pine forest under extensive seasonal drought. Tree Physiol. 2019, 39, 1767–1782.
- Zuromski, L.M.; Bowling, D.R.; Kohler, P.; Frankenberg, C.; Goulden, M.L.; Blanken, P.D.; Lin, J.C. Solar-induced fluorescence detects interannual variation in gross primary production of coniferous forests in the western United States. Geophys. Res. Lett. 2018, 45, 7184–7193.
- Matisons, R.; Krisans, O.; Jansons, A.; Kondratovičs, T.; Elferts, D.; Ievinsh, G. Norway spruce seedlings from an eastern Baltic provenance show tolerance to simulated drought. Forests 2021, 12, 82.
- Konôpková, A.; Húdoková, H.; Ježík, M.; Kurjak, D.; Jamnická, G.; Ditmarová, Ľ.; Gomory, D.; Longauer, R.; Tognetti, R.; Pšidová, E. Origin rather than mild drought stress influenced chlorophyll a fluorescence in contrasting silver fir (Abies alba Mill.) prove-nances. Photosynthetica 2020, 58, 549–559.
- Voltas, J.; Chambel, M.; Prada, A.; Ferrio, J.P. Climate-related variability in carbon and oxygen stable isotopes among popula-tions of Aleppo pine grown in common-garden tests. Trees 2008, 22, 759–769.
- Klein, T.; Matteo, G.; Rotenberg, E.; Cohen, S.; Yakir, D. Differential ecophysiological response of a major Mediterranean pine species across a climatic gradient. Tree Physiol. 2012, 33, 26–36.
- Wang, F.; Zhang, F.; Gou, X.H.; Fonti, P.; Xia, J.Q.; Cao, Z.Y.; Liu, J.G.; Wang, Y.F.; Zhang, J.Z. Seasonal variations in leaf-level photosynthesis and water use efficiency of three isohydric to anisohydric conifers on the Tibetan Plateau. Agric. For. Meteorol. 2021, 308, 108581.
- De Miguel, M.; Cabezas, J.-A.; de María, N.; Sánchez-Gómez, D.; Guevara, M.-Á.; Vélez, M.-D.; Sáez-Laguna, E.; Díaz, L.-M.; Man-cha, J.-A.; Barbero, M.-C.; et al. Genetic control of functional traits related to photosynthesis and water use efficiency in Pinus pi-naster Ait. drought response: Integration of genome annotation, allele association and QTL detection for candidate gene identification. BMC Genom. 2014, 15, 464.
- Farquhar, G.D.; Ehleringer, J.R.; Hubick, K.T. Carbon isotope discrimination and photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 503–537.
- Guy, R.D.; Holowachuk, D.L. Population differences in stable carbon isotope ratio of Pinus contorta Dougl. ex Loud.: Relationship to environment, climate of origin, and growth potential. Can. J. Bot. 2001, 79, 274–283.
- Jensen, A.M.; Eckert, D.; Carter, K.R.; Persson, M.; Warren, J.M. Springtime drought shifts carbon partitioning of recent photo-synthates in 10-year old Picea mariana trees, Causing restricted canopy development. Front. For. Glob. Change 2021, 3, 601046.
- Csilléry, K.; Buchmann, N.; Fady, B. Adaptation to drought is coupled with slow growth, but independent from phenology in marginal silver fir (Abies alba Mill.) populations. Evol. Appl. 2020, 13, 2357–2376.
- Moran, E.; Lauder, J.; Musser, C.; Stathos, A.; Shu, M. The genetics of drought tolerance in conifers. New Phytol. 2017, 216, 1034–1048.
- Fox, H.; Doron-Faigenboim, A.; Kelly, G.; Bourstein, R.; Attia, Z.; Zhou, J.; Moshe, Y.; Moshelion, M.; David-Schwartz, R. Tran-scriptome analysis of Pinus halepensis under drought stress and during recovery. Tree Physiol. 2018, 38, 423–441.
- De María, N.; Guevara, M.Á.; Perdiguero, P.; Vélez, D.; Cabezas, J.A.; López-Hinojosa, M.; Li, Z.; Díaz, L.M.; Pizarro, A.; Mancha, J.; et al. Molecular study of drought response in the Mediterranean conifer Pinus pinaster Ait.: Differential transcriptomic profiling reveals constitutive water deficit-independent drought tolerance mechanisms. Ecol. Evol. 2020, 10, 9788–9807.
- Li, J.; West, J.B.; Hart, A.; Wegrzyn, J.L.; Smith, M.A.; Domec, J.-C.; Loopstra, C.A.; Casola, C. Extensive variation in drought-induced gene expression changes between loblolly pine genotypes. Front. Genet. 2021, 12, 661440.
- Pervaiz, T.; Liu, S.-W.; Uddin, S.; Amjid, M.W.; Niu, S.-H.; Wu, H.X. The transcriptional landscape and hub genes associated with physiological responses to drought stress in Pinus tabuliformis. Int. J. Mol. Sci. 2021, 22, 9604.
- Xiao, F.; Zhao, Y.; Wang, X.R.; Liu, Q.; Ran, J. Transcriptome analysis of needle and root of Pinus massoniana in response to con-tinuous drought stress. Plants 2021, 10, 769.
- Yang, Y.; Hu, X.G.; Zheng, B.; Li, Y.; Wang, T.; Sharma, A.; Yuan, H.W.; Mao, J.F. Transcriptome-wide Identification and Characterization of microRNAs and Their Targets in a Highly Adaptable Conifer Platycladus orientalis. J. Am. Soc. Hortic. Sci. 2022, 147, 7–17.
- Carvalho, A.; Pavia, I.; Fernandes, C.; Pires, J.; Correia, C.; Bacelar, E.; Moutinho-Pereira, J.; Gaspar, M.J.; Bento, J.; Silva, M.E.; et al. Differential physiological and genetic responses of five European Scots pine provenances to induced water stress. J. Plant Physiol. 2017, 215, 100–109.
- Velasco-Conde, T.; Yakovlev, I.; Majada, J.; Aranda, I.; Johnsen, Ø. Dehydrins in maritime pine (Pinus pinaster) and their expres-sion related to drought stress response. Tree Genet. Genomes 2012, 8, 957–973.
- Behringer, D.; Zimmermann, H.; Ziegenhagen, B.; Liepelt, S. Differential gene expression reveals candidate genes for drought stress response in Abies alba (Pinaceae). PLoS ONE 2015, 10, e0124564.
- Li, W.; Lee, J.; Yu, S.; Wang, F.; Lv, W.; Zhang, X.; Li, C.; Yang, J. Characterization and analysis of the transcriptome response to drought in Larix kaempferi using PacBio full-length cDNA sequencing integrated with de novo RNA-seq reads. Planta 2021, 253, 28.
- Du, M.Z.; Ding, G.; Cai, Q.-Z. The transcriptomic responses of Pinus massoniana to drought stress. Forests 2018, 9, 326.
- Lorenz, W.; Alba, R.; Yu, Y.-S.; Bordeaux, J.; Simões, M.; Dean, J. Microarray analysis and scale-free gene networks identify can-didate regulators in drought-stressed roots of loblolly pine (P. taeda L.). BMC Genom. 2011, 12, 264.
- Perdiguero, P.; Collada, C.; Barbero, M.; Casado, G.; Cervera, M.T.; Soto, A. Identification of water stress genes in Pinus pinaster Ait. by controlled progressive stress and suppression-subtractive hybridization. Plant Physiol. Biochem. 2012, 50, 44–53.
- Watkinson, J.; Sioson, A.; Vasquez-Robinet, C.; Shukla, M.; Kumar, D.; Ellis, M.; Heath, L.; Ramakrishnan, N.; Chevone, B.; Wat-son, L.; et al. Photosynthetic acclimation is reflected in specific patterns of gene expression in drought-stressed loblolly pine. Plant Physiol. 2004, 133, 1702–1716.
- Hamanishi, E.; Campbell, M. Genome-wide responses to drought in forest trees. Forestry 2011, 84, 273–283.
- Perdiguero, P.; Barbero, M.; Cervera, M.T.; Soto, A.; Collada, C. Novel conserved segments are associated with differential ex-pression patterns for Pinaceae dehydrins. Planta 2012, 236, 1863–1874.
- Van Ghelder, C.; Parent, G.J.; Rigault, P.; Prunier, J.; Giguere, I.; Caron, S.; Sena, J.S.; Deslauriers, A.; Bousquet, J.; Esmenjaud, D.; et al. The large repertoire of conifer NLR resistance genes includes drought responsive and highly diversified RNLs. Sci. Rep. 2019, 9, 11614.
- Liu, J.J.; Schoettle, A.W.; Sniezko, R.A.; Williams, H.; Zamany, A.; Rancourt, B. Fine dissection of limber pine resistance to Cronartium ribicola using targeted sequencing of the NLR family. BMC Genom. 2021, 22, 567.
- Plomion, C.; Bousquet, J.; Kole, C. Genetics, Genomics and Breeding of Conifers; CRC Press and Edenbridge Science Publishers: New York, NY, USA, 2011; p. 449. ISBN 978-1-57808-719-8.
- Wei, J.; Pei, X.; Hu, X.; Sun, S.; Zhao, C.; Han, R.; Zhao, X. Applications of transcriptome in conifer species. Plant Cell Tissue Organ Cult. 2022, 150, 511–525.
- Cappa, E.P.; Klutsch, J.G.; Sebastian-Azcona, J.; Ratcliffe, B.; Wei, X.; Da Ros, L.; Liu, Y.; Chen, C.; Benowicz, A.; Sadoway, S.; et al. Integrating genomic information and productivity and climate-adaptability traits into a regional white spruce breeding program. PLoS ONE 2022, 17, e0264549.
- Laverdière, J.P.; Lenz, P.; Nadeau, S.; Depardieu, C.; Isabel, N.; Perron, M.; Beaulieu, J.; Bousquet, J. Breeding for adaptation to climate change: Genomic selection for drought response in a white spruce multi-site polycross test. Evol. Appl. 2022, 15, 383–402.
- Mullin, T.; Andersson Gull, B.; Bastien, J.C.; Beaulieu, J.; Burdon, R.; Dvorak, W.; King, J.; Kondo, T.; Krakowski, J.; Lee, S.; et al. Economic importance, breeding objectives and achievements. In Genetics, Genomics and Breeding of Conifers; Plomion, C., Bousquet, J., Kole, C., Eds.; Science Publishers and CRC Press: New York, NY, USA, 2011; pp. 40–127. ISBN 9781578087198.
- Reyer, C.; Brouwers, N.; Rammig, A.; Brook, B.; Epila, J.; Grant, R.F.; Holmgren, M.; Langerwisch, F.; Leuzinger, S.; Lucht, W.; et al. Forest resilience and tipping points at different spatio-temporal scales: Approaches and challenges. J. Ecol. 2015, 103, 5–15.
- Zobel, B.; Talbert, J. Applied Forest Tree Improvement; Blackburn Press: Caldwel, NJ, USA, 2003; ISBN 1930665814.
- Esposito, S.; Carputo, D.; Cardi, T.; Tripodi, P. Applications and trends of machine learning in genomics and phenomics for next-generation breeding. Plants 2019, 9, 34.
- Yao, J.; Sun, D.; Cen, H.; Haixia, X.; Haiyong, W.; Yuan, F. Phenotyping of arabidopsis drought stress response using kinetic chlorophyll fluorescence and multicolor fluorescence imaging. Front. Plant Sci. 2018, 9, 603.
- Lowe, A.; Harrison, N.; French, A. Hyperspectral image analysis techniques for the detection and classification of the early onset of plant disease and stress. Plant Methods 2017, 13, 80.
- Birol, I.; Raymond, A.; Jackman, S.D.; Pleasance, S.; Coope, R.; Taylor, G.A.; Yuen, M.M.; Keeling, C.I.; Brand, D.; Vandervalk, B.P.; et al. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 2013, 29, 1492–1497.
- Nystedt, B.; Street, N.; Wetterbom, A.; Zuccolo, A.; Lin, Y.-C.; Scofield, D.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584.
- Stevens, K.; Wegrzyn, J.; Zimin, A.; Puiu, D.; Crepeau, M.; Cardeno, C.; Paul, R.; Gonzalez-Ibeaz, D.; Koriabine, M.; Holtz-Morris, A.; et al. Sequence of the sugar pine megagenome. Genetics 2016, 204, 1613–1626.
- Zimin, A.; Stevens, K.A.; Crepeau, M.W.; Holtz-Morris, A.; Koriabine, M.; Marçais, G.; Puiu, D.; Roberts, M.; Wegrzyn, J.L.; de Jong, P.J.; et al. Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics 2014, 196, 875–890.
- Scott, A.D.; Zimin, A.V.; Puiu, D.; Workman, R.; Britton, M.; Zaman, S.; Caballero, M.; Read, A.C.; Bogdanove, A.J.; Burns, E.; et al. A reference genome sequence for giant sequoia. G3-Genes Genom. Genet. 2020, 10, 3907–3919.
- Niu, S.H.; Li, J.; Bo, W.H.; Yang, W.F.; Zuccolo, A.; Giacomello, S.; Chen, X.; Han, F.X.; Yang, J.H.; Song, Y.; et al. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell 2022, 185, 204–217.
- Traversari, S.; Giovannelli, A.; Emiliani, G. Wood formation under changing environment: Omics approaches to elucidate the mechanisms driving the early-to-latewood transition in Conifers. Forests 2022, 13, 608.
- Pavy, N.; Lamothe, M.; Pelgas, B.; Gagnon, F.; Birol, I.; Bohlmann, J.; Mackay, J.; Isabel, N.; Bousquet, J. A high-resolution refer-ence genetic map positioning 8.8K genes for the conifer white spruce: Structural genomics implications and correspondence with physical distance. Plant J. 2017, 90, 189–203.
- Bernhardsson, C.; Vidalis, A.; Wang, X.; Scofield, D.G.; Schiffthaler, B.; Baison, J.; Street, N.R.; Garcia-Gil, M.R.; Ingvarsson, P.K. An ultra-dense haploid genetic map for evaluating the highly fragmented genome assembly of Norway spruce (Picea abies). G3-Genes Genom. Genet. 2019, 9, 1623–1632.
- Dong, M.L.; He, Q.W.; Zhao, J.; Zhang, Y.; Yuan, D.S.; Zhang, J.F. Genetic mapping of Prince Rupprecht’s larch (Larix princi-pis-rupprechtii Mayr) by specific-locus amplified fragment sequencing. Genes 2019, 10, 583.
- Jin, Y.Q.; Zhao, W.; Nie, S.; Liu, S.S.; El-Kassaby, Y.A.; Wang, X.R.; Mao, J.F. Genome-wide variant identification and high-density genetic map construction using RADseq for Platycladus orientalis (Cupressaceae). G3-Genes Genom. Genet. 2019, 9, 3663–3672.
- Liu, J.J.; Schoettle, A.W.; Sniezko, R.A.; Yao, F.P.; Zamany, A.; Williams, H.; Rancourt, B. Limber pine (Pinus flexilis James) genetic map constructed by exome-seq provides insight into the evolution of disease resistance and a genomic resource for ge-nomics-based breeding. Plant J. 2019, 98, 745–758.
- Pavy, N.; Gagnon, F.; Rigault, P.; Blais, S.; Deschenes, A.; Boyle, B.; Pelgas, B.; Deslauriers, M.; Clement, S.; Lavigne, P.; et al. Development of high-density SNP genotyping arrays for white spruce (Picea glauca) and transferability to subtropical and nordic congeners. Mol. Ecol. Resour. 2013, 13, 324–336.
- Telfer, E.; Graham, N.; Macdonald, L.; Li, Y.J.; Klapste, J.; Resende, M.; Neves, L.G.; Dungey, H.; Wilcox, P. A high-density exome capture genotype-by-sequencing panel for forestry breeding in Pinus radiata. PLoS ONE 2019, 14, e02222640.
- Nagano, S.; Hirao, T.; Takashima, Y.; Matsushita, M.; Mishima, K.; Takahashi, M.; Iki, T.; Ishiguri, F.; Hiraoka, Y. SNP Genotyping with target amplicon sequencing using a multiplexed primer panel and its application to genomic prediction in Japanese cedar, Cryptomeria japonica(L.f.) D.Don. Forests 2020, 11, 898.
- Bernhardsson, C.; Zan, Y.J.; Chen, Z.Q.; Ingvarsson, P.K.; Wu, H.X. Development of a highly efficient 50K single nucleotide pol-ymorphism genotyping array for the large and complex genome of Norway spruce (Picea abies L. Karst) by whole genome rese-quencing and its transferability to other spruce species. Mol. Ecol. Resour. 2021, 21, 880–896.
- Kastally, C.; Niskanen, A.K.; Perry, A.; Kujala, S.T.; Avia, K.; Cervantes, S.; Haapanen, M.; Kesalahti, R.; Kumpula, T.A.; Mattila, T.M.; et al. Taming the massive genome of Scots pine with PiSy50k, a new genotyping array for conifer research. Plant J. 2022, 109, 1337–1350.
- Muranty, H.; Jorge, V.; Bastien, C.; Lepoittevin, C.; Bouffier, L.; Sanchez, L. Potential for marker-assisted selection for forest tree breeding: Lessons from 20 years of MAS in crops. Tree Genet. Genomes 2014, 10, 1491–1510.
- Grattapaglia, D.; Silva-Junior, O.B.; Resende, R.T.; Cappa, E.P.; Muller, B.S.F.; Tan, B.Y.; Isik, F.; Ratcliffe, B.; El-Kassaby, Y.A. Quantitative genetics and genomics converge to accelerate forest tree breeding. Front. Plant Sci. 2018, 9, 1693.
- Ingvarsson, P.K.; Street, N.R. Association genetics of complex traits in plants. New Phytol. 2011, 189, 909–922.
- Grattapaglia, D. Status and perspectives of genomic selection in forest tree breeding. In Genomic Selection for Crop Improvement: New Molecular Breeding Strategies for Crop Improvement; Varshney, R.K., Roorkiwal, M., Sorrells, M.E., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 199–249.
This entry is offline, you can click here to edit this entry!
