Radiation-induced toxicity to healthy/normal intestinal tissues, especially during radiotherapy, limits the radiation dose necessary to effectively eradicate tumors of the abdomen and pelvis. Although the pathogenesis of intestinal radiation toxicity is highly complex, understanding post-irradiation alterations in protein profiles can provide crucial insights that make radiotherapy safer and more efficient and allow for increasing the radiation dose during cancer treatment. Recent preclinical and clinical studies have advanced our current understanding of the molecular changes associated with radiation-induced intestinal damage by assessing changes in protein expression with mass spectrometry-based approaches and 2-dimensional difference gel electrophoresis. Studies by various groups have demonstrated that proteins that are involved in the inflammatory response, the apoptotic pathway, reactive oxygen species scavenging, and cell proliferation can be targeted to develop effective radiation countermeasures. Moreover, altered protein profiles serve as a crucial biomarkers for intestinal radiation damage. In this review, we present alterations in protein signatures following intestinal radiation damage as detected by proteomics approaches in preclinical and clinical models with the aim of providing a better understanding of how to accomplish intestinal protection against radiation damage.
- radiation
- enteropathy
- proteomics
- intestine
- biomarkers
1. Introduction
Proteomics is a powerful tool to identify and quantitate changes in the expression of thousands of proteins from any tissue, including intestinal tissues, after exposure to any stimuli of external or internal origin. Two proteomics approaches are commonly used to estimate the protein abundance in intestinal tissues. One approach is gel-based and uses either 2-dimensional difference gel electrophoresis (2D-DIGE) or 2-dimensional gel electrophoresis (2-DE), and the other is mass spectrometry (MS). Figure 1 shows a typical flow chart of two different proteomic analysis methods such as gel-based and mass spectrometry-based methods that can be used to assess intestinal radiation toxicity using preclinical and clinical models. These quantitative methods allow the estimation of the relative abundance and identification of the proteome in the intestinal cells in culture and in the intestinal tissue after total body irradiation (TBI) or partial body irradiation (PBI) and in plasma samples after targeted radiation of the abdominal region. These quantitative proteomics approaches continue to provide useful insights into the molecular mechanism of radiation-induced intestinal damage. This review article summarizes the changes in protein profiles in the intestinal cells in culture, intestinal tissues in rodents and nonhuman primates following radiation, or in patients undergoing abdominopelvic radiotherapy. The aim of this review is to identify alterations in the intestinal protein landscape, which may help to target signaling pathways in order to minimize intestinal radiation toxicity.
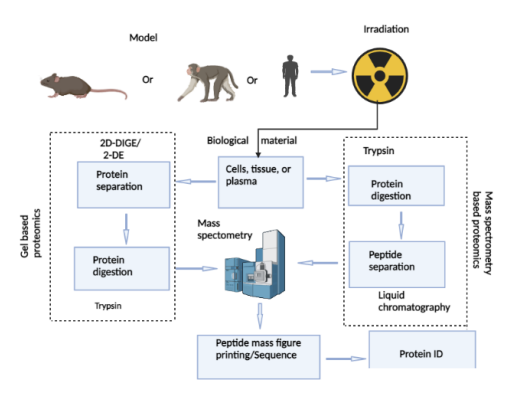
Flow-chart showing the steps involved in intestinal proteomics analysis for gel-based and mass spectrometry-based methods. Briefly, lysates from cells in culture or tissues of rodents and nonhuman primates or plasma from nonhuman primates and humans collected after irradiation are subjected to one of two protein separation methods, gel or mass spectrometry. Subsequently, peptide spectra and corresponding mass to charge ratios are generated by mass spectrometry, searched against protein databases for protein identification and further downstream analysis.
2. Proteomics Methods Used to Separate Proteins
2.1. Gel-Based Proteomics Method
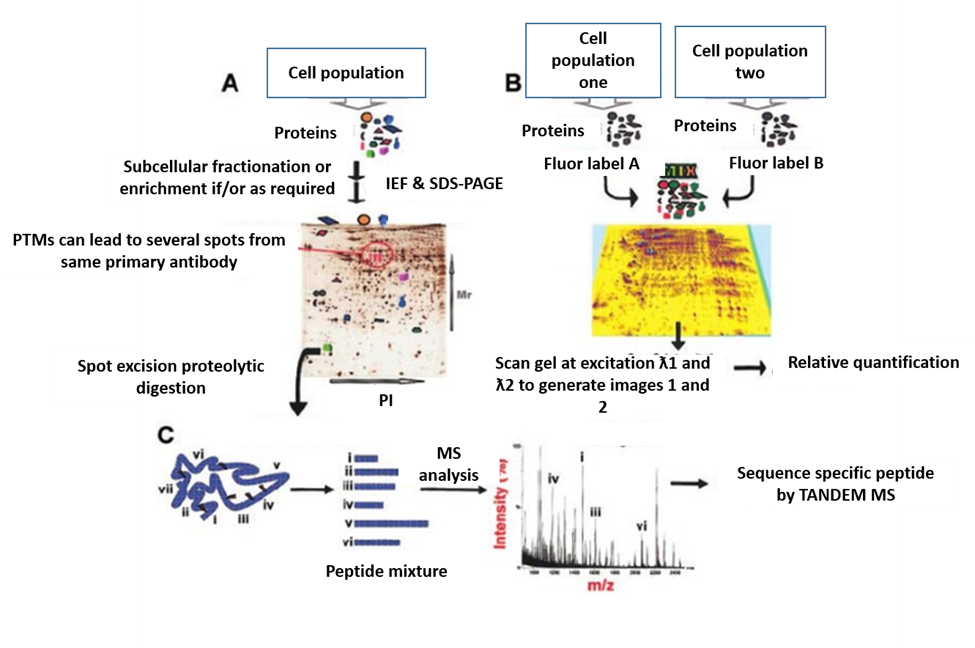
Figure 2. Gel-based proteomics. (A). protein separation by 2-DE; (B). protein separation by 2-D DIGE; and (C). MS for protein identification [10].
2.2. Mass Spectrometry-Based Proteomics
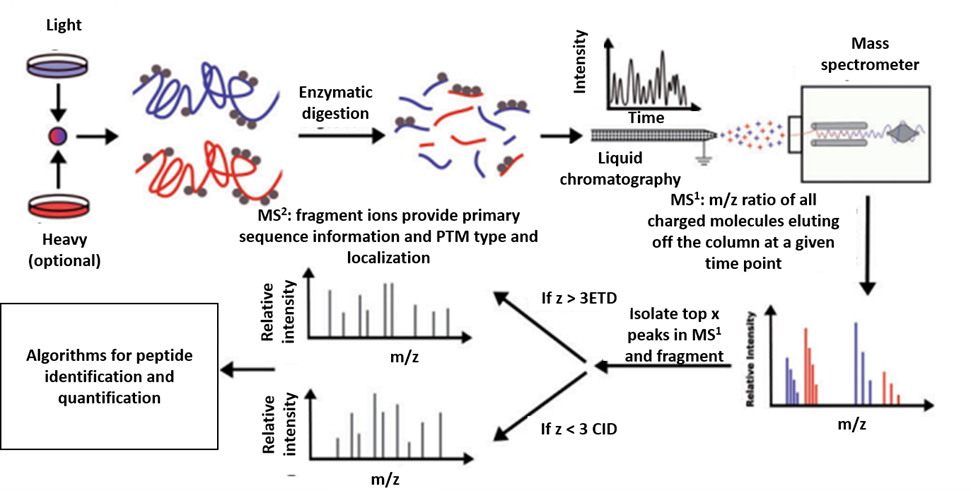
Figure 3. MS-based proteomics. Illustration of the steps involved in MS proteomics analysis [30].
3. Radiation-Induced Change in Protein Profile in Intestinal Cells or Tissues
3.1. Radiation Alters Protein Profiles in Intestinal Cells in Culture
3.2. Radiation Alters Protein Profile in the Intestinal Tissue of Rodents
Proteomics studies of intestinal tissue following irradiation have been extensively investigated, primarily in mice and rats. Table 1 shows preclinical models used for the quantitative proteomics study of radiation damage to normal gastrointestinal tissues and the identification of radiation-responsive proteins. Notably, protein profile changes following irradiation depend on animal strain, sex, and age; radiation dose and type; post-irradiation time interval; and the radiation delivery technique. For example, a proteomics study of intestinal tissues of 6- to 8-week-old male C57BL/6 mice at 1 h after exposure to 9 Gy γ-rays found 17 proteins were expressed only in the irradiated group compared to the unirradiated control group [32]. The dysregulated proteins were involved in biological roles, including post-translational modifications, protein turnover and chaperones, bimolecular transportation and metabolism, cytoskeletal structure, energy production and conversion, and signal transduction mechanisms [32]. Significantly, MYC transcription factor was identified as the only upstream regulator affected by radiation exposure [32]. With the help of commercially available antibody kits, the abnormal expression of ATP synthase subunit D, aldehyde dehydrogenase, Cox5a, CRP, multifaceted C1qbp, Oat, and Pcna was confirmed after irradiation [32]. According to Zhang et al., in comparison to an average of 638 ± 39 protein spots identified by 2-DE in sham-irradiation, exposure to 9 Gy TBI yielded an average of 566 ± 32 protein spots at 3 h and 591 ± 29 at 72 h in the intestinal tissue of 58- to 62-day-old male BALB/c mice [33]. Further analysis by peptide mass fingerprinting revealed that 19 proteins were differentially expressed following irradiation. Proteins involved in redox regulation, such as peroxiredoxin I and glutathione S-transferase P2, were upregulated, while antioxidant protein 2 (also known as 1-Cys peroxiredoxin) was downregulated [33]. Notably, expression of enolase, a glycolytic enzyme, was upregulated in the intestine 3 h after 9 Gy TBI but not at 72 h when compared to sham-irradiated controls [33]. Another study by Lim et al. showed that 1 Gy TBI exposure in 7-week-old C57BL/6 mice resulted in 49 differential intestinal protein spots out of a total of 977 spots analyzed by 2-DE for both irradiated and unirradiated groups [34]. Only 5 of the 49 differential spots were identified and verified by commercially available antibodies [34]. Importantly, the expression of phosphoglycerate kinase 1 (PGK1) was significantly higher at 24 h after 1 Gy TBI in mouse intestinal tissue [34]. Rosen et al. demonstrated that 6- to 8-week-old male CD2F1 mice irradiated with 11 Gy g-rays exhibited 26 differentially altered proteins in the intestine compared to sham-irradiated controls 24 h after exposure [35]; however, the expression of 13 of these proteins was normalized in mice groups treated with either of 2 vitamin E family members, -tocotrienol or tocopherol succinate, prior to radiation exposure [35]. Another intestinal proteomics study conducted in 8- to 10-week-old male C57BL/6J mice exposed to a single uniform X-ray TBI dose of 8, 10, 12, or 14 Gy reported that the expression of 103 proteins was consistently altered 1, 3, and 6 days after exposure [36]. Of these, 46 proteins were consistently activated, and 57 proteins were consistently inhibited [36]. Further analysis showed that due to the consistent alteration in protein expression over the dose range and days, molecular functions associated with thiol ester hydrolase activity, serine-type endopeptidase activity, nucleosomal DNA binding, acyl-CoA hydrolase activity, palmitoyl-CoA hydrolase activity, and carboxylic ester hydrolase activity were significantly elevated, whereas molecular function associated with poly(A) RNA binding was significantly reduced [36]. Furthermore, it was revealed that 16 of the proteins were significantly associated with retinoic acid, including Aldh1a1, Apoa2, Apoe, Rbp2, Rdh7, Ttr, and a series of Akr proteins; 3 proteins were connected to radiation (Eef1d, Ptprc, and Sod2); and 40 proteins were associated with the inflammation [36]. The authors also indicated the expression of 44 upstream regulators was impacted by TBI irradiation [36]. Finally, the authors showed a dose dependent increase in 5 proteins (such as FABP1, FGA, FGB, FGG, and HP) in the intestinal sample of C57BL/6 male mice on day 3 following exposure to 8, 10, 12, and 14 Gy TBI, which could be used as radiation biodosimetric markers for intestinal injury [36]. Han et al. analyzed the intestinal proteome of 6-week-old female C57BL/6J mice exposed to 7 Gy g-rays TBI 10 days after irradiation and identified 186 proteins in the control group, 270 proteins in the irradiated group, and 238 proteins in a group irradiated and treated with human placenta-derived stem cells (hPDSCs) [37]. Further analysis revealed 68 uniquely expressed proteins in the irradiated only mice compared to 27 in the control group and 38 in the radiation plus hPDSCs-treated group [37]. IL-10, glycoprotein, TIMP-1, and antioxidant protein, GST mu type 1, which decreased in expression following radiation exposure, were verified by immunoblot; however, the administration of hPDSCs increased the expression of these proteins [37]. Moreover, Wang et al. examined intestinal tissue of male C57BL/6J mice 3 days after 13 Gy 137Cs abdominal g-ray irradiation and identified 1,279 proteins that were differentially expressed compared to sham-irradiated controls [38].
Two proteomics studies of intestinal tissues have utilized abdominal irradiation of male Sprague–Dawley rats. In one of these studies, intestinal tissues of adult rats were examined 4 days after exposure to 10 Gy g-rays, revealing 86 differentially expressed proteins involved in lipid, protein, carbohydrate, and other metabolic and cellular processes [39]. In the second study, 6-week-old rats were exposed to 20 Gy X-rays, and 10 weeks after exposure, 6,692 proteins were identified, of which 5,756 were quantified [40]. Of the quantified proteins, 320 were significantly altered [40]. The differentially expressed proteins were involved primarily in biological regulation and single-organism processes, metabolic processes, and the response to stimulus [40]. Validation by parallel reaction monitoring showed that proteins associated with complement and coagulation cascades (FGG, C3, and F2), regulation of the actin cytoskeleton (F2, ITGB2, and ITGAM), and leukocyte trans-endothelial migration (CYBB, ITGB2, and ITGAM) were upregulated [40]. The findings of these studies establish the fact that exposure to ionizing radiation alters the intestinal protein expression profile.
Table 1. Preclinical rodent models used for quantitative proteomics study in the intestinal tissue following radiation exposure.
|
Strain |
Sex |
Age |
Tissue Type |
Radiation Dose (Gy) |
Tissue Harvest Time |
Mode of Radiation |
Radiation Type |
Radiation-Responsive Proteins |
Ref |
|
C57BL/6 mice |
Male |
6–8 weeks |
Jejunum |
9 |
1 h |
TBI |
g-rays |
ATP synthase subunit D, aldehyde dehydrogenase, Cox 5a, CRP, multifaceted C1qbp, Oat, Pcna |
|
|
Bal b/c mice |
Male |
58–62 days |
Intestinal epithelia |
9 |
3 and 72 h |
TBI |
g-rays |
Enolase and peroxiredoxin |
[33] |
|
C57BL/6 mice |
Female |
7 weeks |
|
1 |
24 h |
TBI |
g-rays |
Phosphoglycerate kinase 1 |
[34] |
|
CD2F1 mice |
Male |
6–8 weeks |
Jejunum |
11 |
24 h |
TBI |
g-rays |
Cytoplasmic actin 2, dihydropyrimidinaserelated protein 2, ezrin, elongation factor 2, plastin-1, and peroxiredoxin-1 |
[35] |
|
C57BL/6J mice |
Male |
8–10 weeks |
Ileum |
8, 10, 12 and 14 |
1, 3, and 6 days |
TBI |
X-rays |
Ctsc, Cen pv, amy2, DUOX2, Try4, Fabp1, Dsp |
[36] |
|
C57BL/6J mice |
Female |
6 weeks |
Small intestines |
7 |
10 days |
TBI |
g-rays |
IL-10, glycoprotein, TIMP-1, and antioxidant protein, GST mu type 1 |
[37] |
|
C57BL/6J mice |
Male |
6–8 weeks |
Small intestines |
13 |
3 days |
ABI |
g-rays |
Gpx3, Sod3 |
[38] |
|
Sprague–Dawley rats |
Male |
Adult |
Ileum |
10 |
4 days |
ABI |
g-rays |
Gelsolin, Prelamin-A/C, |
[39] |
|
Sprague–Dawley rats |
Male |
6 weeks |
Large intestine |
20 |
10 weeks |
ABI |
X-rays |
FGG, THBS1, AGT, F2, C3, ITGAM, ITGB2, CYBB, QSOX1 |
[40] |
3.3. Radiation Alters Protein Profile in the Intestinal Tissue of Nonhuman Primates
4. Therapeutic Radiation Alters the Plasma Protein Profile of Rectal Cancer Patients
5. Signaling Pathways Altered as a Result of Intestinal Radiation Toxicity
In addition, radiation was shown to affect various signaling pathways in nonhuman primates. For example, 4 canonical pathways—GP6 signaling pathway, acute phase response signaling, LXR/RXR activation, and intrinsic prothrombin activation pathway—were dysregulated in the intestinal tissue of nonhuman primates following 12 Gy PBI/2.5 BM at various time points ranging from 4 to 22 days [41]. Table 2 shows different pathways affected in intestinal tissues following irradiation.
Table 2. Pathways dysregulated by radiation exposure in normal intestinal tissues.
|
Strain |
Age |
Tissue type |
Radiation dose (Gy) |
Post-irradiation interval |
Mode of Radiation |
Radiation types |
Radiation-impacted pathways |
Ref |
|
C57BL/6 mice |
6-8 weeks |
Jejunum |
9 |
1 h |
TBI |
g-rays |
Proteasome and protein processing in the endoplasmic reticulum |
|
|
CD2F1 mice |
6-8 weeks |
Jejunum |
11 |
24 h |
TBI |
g-rays |
Rho family GTPases Signaling, glycolysis I, xenobiotic metabolism, 14-3-3-mediated signaling, and retinol biosynthesis |
[35] |
|
C57BL/6J mice |
8-10 weeks |
Ileum |
8, 10, 12 and 14 |
1, 3, and 6 days |
TBI |
X-rays |
Protein kinase A, acute phase response, and LXR/RXR signaling |
[36] |
|
C57BL/6J mice |
6-8 weeks |
Small intestines |
13 |
3 days |
ABI |
g-rays |
DNA damage and apoptosis signaling |
[38] |
|
Sprague–Dawley rats |
Adult |
Ileum |
10 |
4 days |
ABI |
g-rays |
FAS and glycolysis signaling pathway |
[39] |
|
Sprague–Dawley rats |
6 weeks |
Large intestines |
20 |
10 weeks |
ABI |
X-rays |
Complement and coagulation cascades, amoebiasis, phagosome, lysosome, focal adhesion, proteoglycans in cancer, and oxytocin signaling |
[40] |
|
Nonhuman primate (Macaca mulatta) |
Adult |
Jejunum |
12 |
4, 8/9, 11/12, 15, and 21/22 days |
PBI |
X-rays |
GP6 Signaling Pathway, acute phase response signaling, LXR/RXR pathway, and intrinsic prothrombin activation pathway |
[41] |
6. Discussion
In this review, we have discussed the importance of studying intestinal proteomics in regard to radiation toxicity, provided an overview of methods used for proteomics studies, and summarized the total number of altered proteins following intestinal radiation injury as well as the major intestinal pathways dysregulated after radiation damage. Studies of intestinal proteomics following radiation injury may help to identify crucial protein biomarkers that can be targeted to develop new countermeasure strategies to limit acute and delayed intestinal toxicity. Limiting intestinal radiation toxicity would not only improve the quality of life of radiation victims or radiation-treated cancer survivors who are suffering from intestinal toxicity, but it also would significantly reduce the burden on health care systems.
Identifying protein biomarkers by proteomics approaches is a challenging task as protein profiles change based on a number of factors, including animal strain, age, species, and sex; radiation dose, quality and mode of delivery; and post-irradiation time points. For example, 17 proteins were differentially expressed in male C57BL/6 mice after 1 h of 9 Gy TBI with g-rays, whereas 49 proteins were differentially expressed in female C57BL/6 mice 24 h after 1 Gy TBI with g-rays as compared to sham-irradiated control groups [32,34], suggesting radiation dose, animal sex, and post-irradiation times are confounding factors in altering protein level in the intestinal tissue. Moreover, a comparison of 2 other intestinal proteomics studies, where the radiation delivery technique (TBI) and radiation type (g-rays) were the same, revealed that a total of 26 proteins were altered in male CD2F1 mice 24 h after 11 Gy irradiation, while 95 proteins were differentially expressed 10 days after 7 Gy irradiation in female C57BL/6 J mice compared to the respective sham-irradiated controls [35,37]. Similarly, in Sprague–Dawley rats, 86 intestinal proteins were altered on day 4 after 10 Gy g-ray, and 320 intestinal proteins were altered at 10 weeks after 20 Gy X-ray irradiation [39,41]. These results further confirm the challenges faced in identifying biomarkers in proteomics studies. However, with the advancement of protein separation techniques, the proteins in a sample of interest can be characterized more accurately.
Because it is not practical to collect intestinal samples for proteomics studies from patients accidentally exposed to radiation or patients with cancer being treated with radiation, identifying protein biomarkers by systematically scanning the changes in protein profiles from plasma samples may serve as an alternative approach for predicting the extent of intestinal radiation damage. Plasma collection is relatively convenient and is a less invasive method. A number of studies have shown that after intestinal radiation damage, the expression of the same proteins altered in the plasma sample as well as in the intestinal tissues, suggesting plasma proteins may serve as candidate biomarkers of radiation toxicity to the gastrointestinal tract. For example, serum amyloid A1 SSA1 and c-reactive protein are significantly altered both in plasma samples and intestinal tissues of nonhuman primates following 12 Gy PBI [36,43]. Interestingly, the same heat shock protein, amyloid A1 SSA1, was also altered in the plasma of patients with rectal cancer following 5 fractions of 5 Gy radiation for 5 consecutive days [42]. These findings may provide the impetus for more in-depth research in order to identify plasma protein biomarkers to assess intestinal radiation damage.
All proteins directly or indirectly influence one or more signaling pathways. Therefore, the altered intestinal protein profile following irradiation can be used to predict which pathways are dysregulated. Targeting dysregulated pathways may counteract intestinal radiation injury. In this regard, identifying a common pathway that is dysregulated irrespective of radiation dose and quality, post-irradiation time, and species would be an invaluable target for protection from intestinal radiation damage. Studies have shown that acute phase response signaling and the LXR/RXR pathway are dysregulated both in C57BL/6J following exposure to various doses of TBI with X-rays (8, 10, 12, or 14 Gy) and in rhesus macaques following exposure to 12 Gy PBI with X-rays [36,41]. Similarly, the glycolysis signaling pathway was dysregulated in the intestinal tissue of C57BL/6J exposed to 13 Gy ABI with 137Cs g-rays and in Sprague–Dawley rats exposed to 10 Gy ABI with 60Co g-rays [38,39]. Finally, the coagulation cascade was dysregulated in the intestinal tissue of Sprague–Dawley rats after exposure to 20 Gy ABI with X-rays and in plasma samples of rhesus macaques after exposure to 12 Gy PBI with a 6 MV linear accelerator [40,43].
When the experimental settings (in terms animal species and strains, absorbed dose and dose rate, tissue harvest times, and radiation delivery techniques) are different, causal connections between radiation-induced tissue damage and pathological changes based on proteomics data are particularly difficult to establish. However, the cited literature in this review article indicates that acute effects of radiation cause apoptosis [38], mitochondrial dysfunction [32,37], and altered cell signaling [35]. Radiation causes damage to the mucosal epithelial layer by inducing apoptotic signaling pathways, thereby increasing the risk of luminal microbe infiltration into the circulatory system, resulting in sepsis if treatment does not happen in a timely manner. In addition, radiation downregulates expression of proteins involved in normal mitochondrial activity, such as ATP synthase sub-unit D, Cox5a and antioxidant protein 2 [32,37]. The compromise of mitochondrial activity facilitates generation of excessive reactive oxygen species, leading to oxidative damage of functional activity of several proteins, namely proteasomes [32]. Proteasomes remove damaged proteins which result from radiation-induced tissue damage. To restore mitochondrial function and counteract the excessive ROS generation, glycolytic enzymes (such as enolase and phosphoglycerate kinase 1) [33,34], and ROS scavenging enzymes (such as peroxiredoxin I and glutathione S-transferase P2) [33,35] may be increased, respectively. In addition, radiation inhibits G-protein coupled receptors (GPCRs) from transmission of cell signals because of impairment of structural proteins (such as cytoplasmic actin 2, lamin B1, and ezrin) [35,39], which can adversely affect several cell signaling pathways, including those that play a crucial role in radiation protection. For example, radiation suppresses GPCR-mediated retinoic pathway [35]. A previous study has shown that retinoic acid and TIMP1 prevent radiation-induced apoptosis in endothelial cells [44]. Apoptosis, mitochondrial dysfunction, and altered cell signaling trigger activation of immune cells leading to persistent systemic inflammation. These activated immune cells interact with endothelial cells that form the inner layer of the blood vasculature and transmigrate through the endothelial layer to reach to the damage tissues [40]. Persistent inflammation drives delayed effects of radiation by activation of the complement cascade, enhancing coagulopathy, and fueling thrombotic changes [40,41]. Figure 4 summarizes events and resulting pathological conditions following radiation at both cellular and tissue levels.
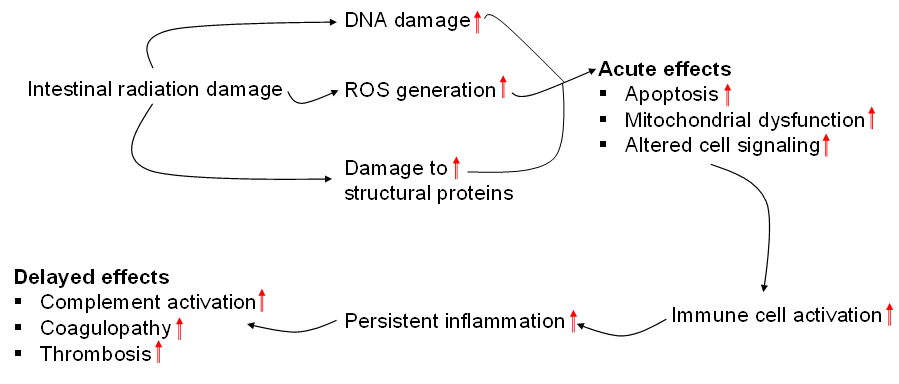
Figure 4. Summary of events and resulting pathological condition because of acute and delayed effects of intestinal radiation injury.
In conclusion, data from comprehensive radiation proteomics in the intestinal tissue have deepened the knowledge of molecular mechanisms involved in intestinal radiation toxicity, which holds great promise but needs further validation. Further research and state-of-the-art proteomics have the potential to identify new diagnostic and therapeutic markers to limit radiation-induced intestinal toxicity. In addition, determining the signaling pathways associated with early and delayed reactions of radiation will provide further insight regarding the management of intestinal toxicity.
This entry is adapted from the peer-reviewed paper 10.3390/genes13112006
