Protein S-glutathionylation (SSG) is a reversible post-translational modification (PTM) featuring the conjugation of glutathione to a protein cysteine thiol. SSG can alter protein structure, activity, subcellular localization, and interaction with small molecules and other proteins. Thus, it plays a critical role in redox signaling and regulation in various physiological activities and pathological events. Many approaches have been developed for the detection of SSG, including direct detection, selective reduction and tagging approaches, and chemoselective probe-based approaches. Utilization of these methods in profiling the SSG proteome had been reported in various biological systems.
- redox
- redox proteomics
- post-translational modification
1. Direct Detection
2. Selective Reduction and Tagging Approaches
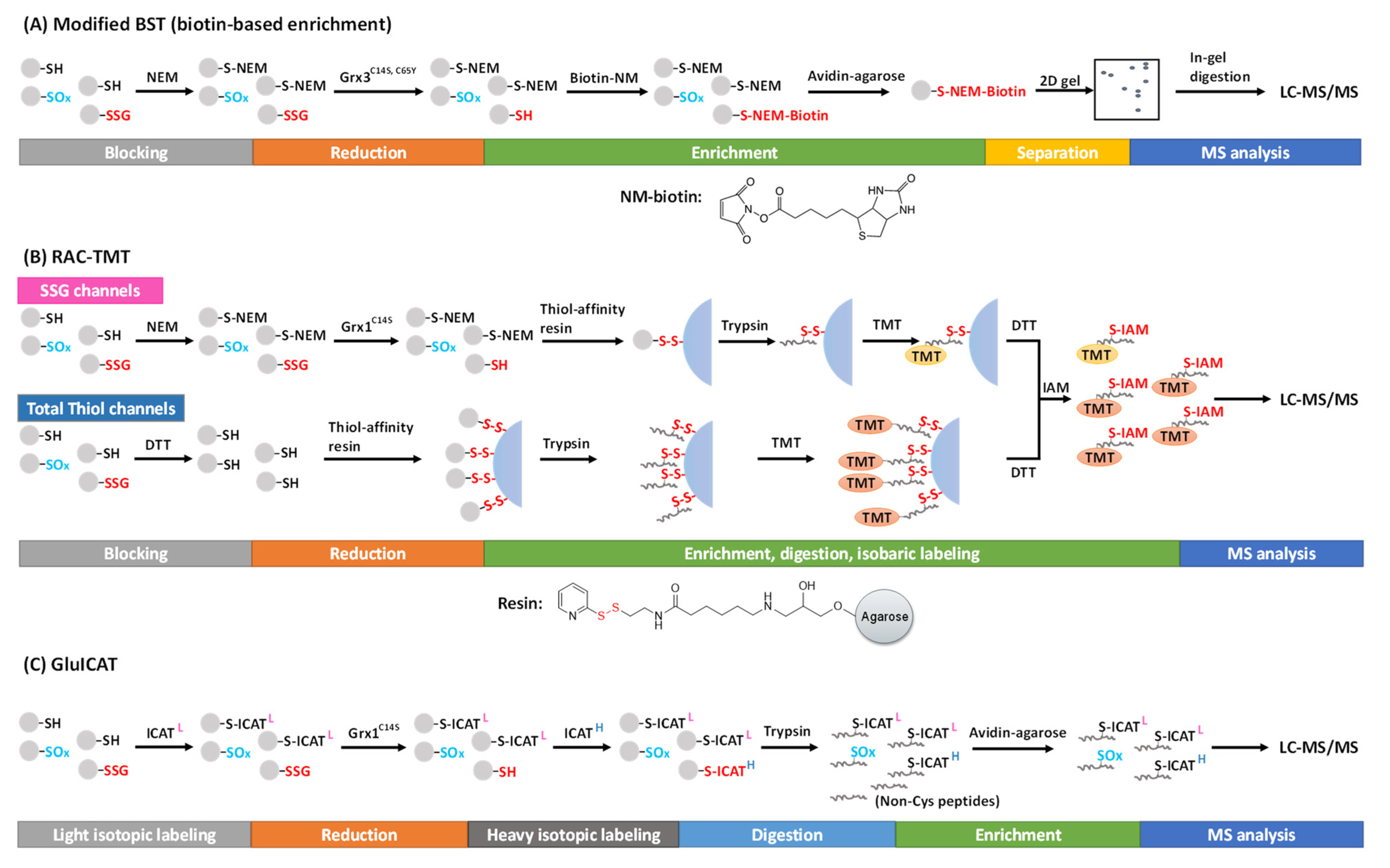
3. Chemoselective Probes-Based Approaches
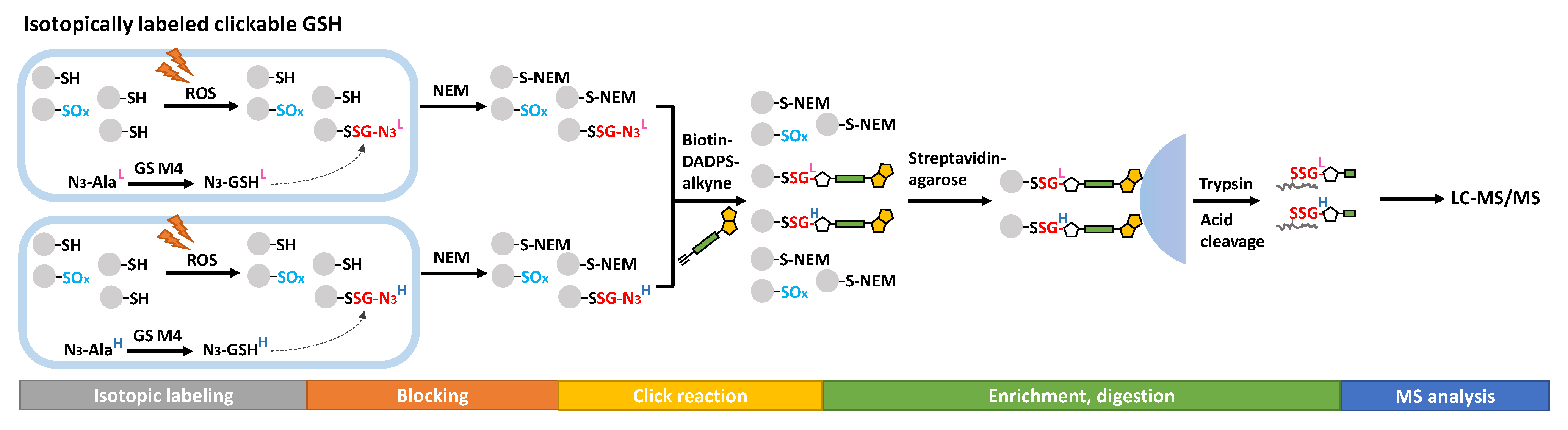
4. Global Profiling of the SSG Proteome
| Biological System | Method | Coverage of the SSG Proteome | Experiment Condition | Significance | Ref. |
|---|---|---|---|---|---|
| Mouse lung | RAC-TMT | ~7600 SSG sites. | Hyperoxia vs. basal; Neonatal wild-type vs. overexpression (β-ENaC) |
Landscape view of SSG-modified proteome in mouse lung and the impact of hypoxia on the SSG proteome | [30] |
| RAW 264.7 mouse macrophages | RAC-TMT | Occupancy for 4099 SSG sites | Basal condition | Proteome-wide quantification of both SSG and total oxidation occupancy under physiological conditions, revealing cellular compartmentation of redox homeostasis. | [25] |
| Mouse skeletal muscle | RAC-TMT | Occupancy for >2200 SSG sites and changes due to muscle fatiguing | Gastrocnemius muscle with and without fatiguing contractions | Increased muscle protein SSGs identified following a single bout of fatiguing contraction | [24] |
| HL-1 cardiomyocyte | Clickable GSH | 1398 SSG-peptides in isotopic duplex experiment | H2O2 treatment (1 mM) | In vivo isotopic tagging of protein SSGs for direct enrichment and quantitative detection; Validation (by Western blot and site mutation) of two structural proteins of interest. | [43] |
| Mouse liver | Modified RAC-TMT | 724 SSG-modified proteins | Basal condition, GSTP-nulled mice | The SSG proteome mediated enzymatically by GSTP | [28] |
| Synechocystis sp. PCC6803 | GSSG-Biotin | 349 proteins with SSG (protein level enrichment); 145 SSG sites (peptide level enrichment) |
Lysate treated with GSSG-biotin | First SSG proteome profiling in cyanobacteria by GSSG-biotin and LC-MS/MS. | [44] |
| Streptococcus mutans UA159 | IodoTMT switch strategy | 357 SSG sites | Wild-type vs. mutants | SSG profiling in in bacteria and mutants; Site mutagenesis validation for SSG function on a thioredoxin-like protein. | [45] |
| Human skin fibroblasts | GluICAT | 2307 SSG sites | LHON patients vs. healthy controls | Quantify the ratio of SSG and free thiols with heavy or light ICAT | [32] |
This entry is adapted from the peer-reviewed paper 10.3390/antiox11112272
References
- Casagrande, S.; Bonetto, V.; Fratelli, M.; Gianazza, E.; Eberini, I.; Massignan, T.; Salmona, M.; Chang, G.; Holmgren, A.; Ghezzi, P. Glutathionylation of human thioredoxin: A possible crosstalk between the glutathione and thioredoxin systems. Proc. Natl. Acad. Sci. USA 2002, 99, 9745–9749.
- Michelet, L.; Zaffagnini, M.; Vanacker, H.; Le Maréchal, P.; Marchand, C.; Schroda, M.; Lemaire, S.D.; Decottignies, P. In Vivo Targets of S-Thiolation in Chlamydomonas reinhardtii. J.Bio. Chem. 2008, 283, 21571–21578.
- Fratelli, M.; Demol, H.; Puype, M.; Casagrande, S.; Villa, P.; Eberini, I.; Vandekerckhove, J.; Gianazza, E.; Ghezzi, P. Identification of proteins undergoing glutathionylation in oxidatively stressed hepatocytes and hepatoma cells. Proteomics 2003, 3, 1154–1161.
- Schuppe-Koistinen, I.; Moldeus, P.; Bergman, T.; Cotgreave, I.A. S-Thiolation of human endothelial cell glyceraldehyde-3-phosphate dehydrogenase after hydrogen peroxide treatment. Eur. J. Biochem. 1994, 221, 1033–1037.
- Rokutan, K.; Johnston, R.B.; Kawai, K. Oxidative stress induces S-thiolation of specific proteins in cultured gastric mucosal cells. Am. J. Physiol.-Gastrointest. Liver Physiol. 1994, 266, G247–G254.
- Ward, N.E.; Stewart, J.R.; Ioannides, C.G.; O’Brian, C.A. Oxidant-Induced S-Glutathiolation Inactivates Protein Kinase C-α (PKC-α): A Potential Mechanism of PKC Isozyme Regulation. Biochemistry 2000, 39, 10319–10329.
- Shenton, D.; Grant, C.M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003, 374, 513–519.
- Dalle-Donne, I.; Giustarini, D.; Colombo, R.; Milzani, A.; Rossi, R. S-glutathionylation in human platelets by a thiol-disulfide exchange-independent mechanism. Free Radic. Biol. Med. 2005, 38, 1501–1510.
- Fratelli, M.; Gianazza, E.; Ghezzi, P. Redox proteomics: Identification and functional role of glutathionylated proteins. Expert Rev. Proteom. 2004, 1, 365–376.
- Domenico, F.D.; Cenini, G.; Sultana, R.; Perluigi, M.; Uberti, D.; Memo, M.; Butterfield, D.A. Glutathionylation of the Pro-apoptotic Protein p53 in Alzheimer’s Disease Brain: Implications for AD Pathogenesis. Neurochem. Res. 2009, 34, 727–733.
- Kil, I.S.; Kim, S.Y.; Park, J.-W. Glutathionylation regulates IκB. Biochem. Biophys. Res. Commun. 2008, 373, 169–173.
- Carvalho, A.N.; Marques, C.; Guedes, R.C.; Castro-Caldas, M.; Rodrigues, E.; van Horssen, J.; Gama, M.J. S-Glutathionylation of Keap1: A new role for glutathione S-transferase pi in neuronal protection. FEBS Lett. 2016, 590, 1455–1466.
- Jin, Y.; Diffee, G.M.; Colman, R.J.; Anderson, R.M.; Ge, Y. Top-down Mass Spectrometry of Sarcomeric Protein Post-translational Modifications from Non-human Primate Skeletal Muscle. J. Am. Soc. Mass Spectrom. 2019, 30, 2460–2469.
- Wei, L.; Gregorich, Z.R.; Lin, Z.; Cai, W.; Jin, Y.; McKiernan, S.H.; McIlwain, S.; Aiken, J.M.; Moss, R.L.; Diffee, G.M.; et al. Novel Sarcopenia-related Alterations in Sarcomeric Protein Post-translational Modifications (PTMs) in Skeletal Muscles Identified by Top-down Proteomics. Mol. Cell. Proteom. 2018, 17, 134–145.
- Li, X.; Day, N.J.; Feng, S.; Gaffrey, M.J.; Lin, T.D.; Paurus, V.L.; Monroe, M.E.; Moore, R.J.; Yang, B.; Xian, M.; et al. Mass spectrometry-based direct detection of multiple types of protein thiol modifications in pancreatic beta cells under endoplasmic reticulum stress. Redox Biol. 2021, 46, 102111.
- Jaffrey, S.R.; Snyder, S.H. The Biotin Switch Method for the Detection of S-Nitrosylated Proteins. Sci. STKE 2001, 2001, pl1.
- Lind, C.; Gerdes, R.; Hamnell, Y.; Schuppe-Koistinen, I.; von Löwenhielm, H.B.; Holmgren, A.; Cotgreave, I.A. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch. Biochem. Biophys. 2002, 406, 229–240.
- Su, D.; Gaffrey, M.J.; Guo, J.; Hatchell, K.E.; Chu, R.K.; Clauss, T.R.W.; Aldrich, J.T.; Wu, S.; Purvine, S.; Camp, D.G.; et al. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radic. Biol. Med. 2014, 67, 460–470.
- Bushweller, J.H.; Aaslund, F.; Wuethrich, K.; Holmgren, A. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14.fwdarw.S) and its mixed disulfide with glutathione. Biochemistry 1992, 31, 9288–9293.
- Reynaert, N.L.; Ckless, K.; Guala, A.S.; Wouters, E.F.M.; van der Vliet, A.; Janssen-Heininger, Y.M.W. In situ detection of S-glutathionylated proteins following glutaredoxin-1 catalyzed cysteine derivatization. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2006, 1760, 380–387.
- Guo, J.; Gaffrey, M.J.; Su, D.; Liu, T.; Camp, D.G., II; Smith, R.D.; Qian, W.J. Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat. Protoc. 2014, 9, 64–75.
- Liu, T.; Qian, W.-J.; Strittmatter, E.F.; Camp, D.G.; Anderson, G.A.; Thrall, B.D.; Smith, R.D. High-Throughput Comparative Proteome Analysis Using a Quantitative Cysteinyl-peptide Enrichment Technology. Anal. Chem. 2004, 76, 5345–5353.
- Duan, J.; Kodali, V.K.; Gaffrey, M.J.; Guo, J.; Chu, R.K.; Camp, D.G.; Smith, R.D.; Thrall, B.D.; Qian, W.J. Quantitative Profiling of Protein S-Glutathionylation Reveals Redox-Dependent Regulation of Macrophage Function during Nanoparticle-Induced Oxidative Stress. ACS Nano 2016, 10, 524–538.
- Kramer, P.A.; Duan, J.; Gaffrey, M.J.; Shukla, A.K.; Wang, L.; Bammler, T.K.; Qian, W.J.; Marcinek, D.J. Fatiguing contractions increase protein S-glutathionylation occupancy in mouse skeletal muscle. Redox Biol. 2018, 17, 367–376.
- Duan, J.; Zhang, T.; Gaffrey, M.J.; Weitz, K.K.; Moore, R.J.; Li, X.; Xian, M.; Thrall, B.D.; Qian, W.J. Stochiometric quantification of the thiol redox proteome of macrophages reveals subcellular compartmentalization and susceptibility to oxidative perturbations. Redox Biol. 2020, 36, 101649.
- Zhang, T.; Gaffrey, M.J.; Li, X.; Qian, W.J. Characterization of cellular oxidative stress response by stoichiometric redox proteomics. Am. J. Physiol. Cell Physiol. 2021, 320, C182–C194.
- Day, N.J.; Gaffrey, M.J.; Qian, W.J. Stoichiometric Thiol Redox Proteomics for Quantifying Cellular Responses to Perturbations. Antioxidants 2021, 10, 499.
- McGarry, D.J.; Chen, W.; Chakravarty, P.; Lamont, D.L.; Wolf, C.R.; Henderson, C.J. Proteome-wide identification and quantification of S-glutathionylation targets in mouse liver. Biochem. J. 2015, 469, 25–32.
- Campbell, M.D.; Duan, J.; Samuelson, A.T.; Gaffrey, M.J.; Merrihew, G.E.; Egertson, J.D.; Wang, L.; Bammler, T.K.; Moore, R.J.; White, C.C.; et al. Improving mitochondrial function with SS-31 reverses age-related redox stress and improves exercise tolerance in aged mice. Free Radic. Biol. Med. 2019, 134, 268–281.
- Zhang, T.; Day, N.J.; Gaffrey, M.; Weitz, K.K.; Attah, K.; Mimche, P.N.; Paine, R., III; Qian, W.J.; Helms, M.N. Regulation of hyperoxia-induced neonatal lung injury via post-translational cysteine redox modifications. Redox Biol. 2022, 55, 102405.
- Leichert, L.I.; Gehrke, F.; Gudiseva, H.V.; Blackwell, T.; Ilbert, M.; Walker, A.K.; Strahler, J.R.; Andrews, P.C.; Jakob, U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 8197–8202.
- Zhou, L.; Chan, J.C.Y.; Chupin, S.; Gueguen, N.; Desquiret-Dumas, V.; Koh, S.K.; Li, J.; Gao, Y.; Deng, L.; Verma, C.; et al. Increased Protein S-Glutathionylation in Leber’s Hereditary Optic Neuropathy (LHON). Int. J. Mol. Sci. 2020, 21, 3027.
- Chan, J.C.Y.; Soh, A.C.K.; Kioh, D.Y.Q.; Li, J.; Verma, C.; Koh, S.K.; Beuerman, R.W.; Zhou, L.; Chan, E.C.Y. Reactive Metabolite-induced Protein Glutathionylation: A Potentially Novel Mechanism Underlying Acetaminophen Hepatotoxicity. Mol. Cell. Proteom. 2018, 17, 2034–2050.
- Xiao, H.; Jedrychowski, M.P.; Schweppe, D.K.; Huttlin, E.L.; Yu, Q.; Heppner, D.E.; Li, J.; Long, J.; Mills, E.L.; Szpyt, J.; et al. A Quantitative Tissue-Specific Landscape of Protein Redox Regulation during Aging. Cell 2020, 180, 968–983.e24.
- Shakir, S.; Vinh, J.; Chiappetta, G. Quantitative analysis of the cysteine redoxome by iodoacetyl tandem mass tags. Anal. Bioanal. Chem. 2017, 409, 3821–3830.
- Demasi, M.; Silva, G.M.; Netto, L.E. 20 S proteasome from Saccharomyces cerevisiae is responsive to redox modifications and is S-glutathionylated. J. Biol. Chem. 2003, 278, 679–685.
- Dixon, D.P.; Skipsey, M.; Grundy, N.M.; Edwards, R. Stress-Induced Protein S-Glutathionylation in Arabidopsis. Plant Physiol. 2005, 138, 2233–2244.
- Brennan, J.P.; Miller, J.I.A.; Fuller, W.; Wait, R.; Begum, S.; Dunn, M.J.; Eaton, P. The Utility of N,N-Biotinyl Glutathione Disulfide in the Study of Protein S-Glutathiolation. Mol. Cell. Proteom. 2006, 5, 215–225.
- Sullivan, D.M.; Wehr, N.B.; Fergusson, M.M.; Levine, R.L.; Finkel, T. Identification of Oxidant-Sensitive Proteins: TNF-α Induces Protein Glutathiolation. Biochemistry 2000, 39, 11121–11128.
- Samarasinghe, K.T.G.; Munkanatta Godage, D.N.P.; Zhou, Y.; Ndombera, F.T.; Weerapana, E.; Ahn, Y.-H. A clickable glutathione approach for identification of protein glutathionylation in response to glucose metabolism. Mol. BioSystems 2016, 12, 2471–2480.
- Kekulandara, D.N.; Samarasinghe, K.T.G.; Godage, D.N.P.M.; Ahn, Y.-H. Clickable glutathione using tetrazine-alkene bioorthogonal chemistry for detecting protein glutathionylation. Org. Biomol. Chem. 2016, 14, 10886–10893.
- VanHecke, G.C.; Abeywardana, M.Y.; Ahn, Y.-H. Proteomic Identification of Protein Glutathionylation in Cardiomyocytes. J. Proteome Res. 2019, 18, 1806–1818.
- VanHecke, G.C.; Yapa Abeywardana, M.; Huang, B.; Ahn, Y.-H. Isotopically Labeled Clickable Glutathione to Quantify Protein S-Glutathionylation. ChemBioChem 2020, 21, 853–859.
- Chardonnet, S.; Sakr, S.; Cassier-Chauvat, C.; Le Maréchal, P.; Chauvat, F.; Lemaire, S.D.; Decottignies, P. First Proteomic Study of S-Glutathionylation in Cyanobacteria. J. Proteome Res. 2015, 14, 59–71.
- Li, Z.; Zhang, C.; Li, C.; Zhou, J.; Xu, X.; Peng, X.; Zhou, X. S-glutathionylation proteome profiling reveals a crucial role of a thioredoxin-like protein in interspecies competition and cariogenecity of Streptococcus mutans. PLOS Pathog. 2020, 16, e1008774.
