Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biology
Optimizing the approaches and compounds utilized to manage plant pests/pathogens via more reliable and safer techniques represents a current pressing challenge for sustainable agricultural systems. The dissatisfaction with synthesized pesticides, such as chemical nematicides, has been increasing. These trends concerning a lack of content for numerous pesticides are emanating from their unfavorable impacts, e.g., their unacceptable influences on ecological contamination and human health, non-target beneficial organisms, and the development of resistance-breaking pathotypes.
- nematode management
- biocontrol
- mechanisms
- plant–nematode interactions
1. Role of Sampling and Extraction Methods in Grasping Plant–Nematode Interactions
Optimizing sampling and extraction methods for PPNs is fundamental for obtaining a sound understanding of these interactions and exploiting them in order to upgrade benign and reliable nematode control. Because sampling is closely associated with the study of every plant–nematode interaction and consequent PPN management, its possible flaws will negatively affect all relevant scopes. Basically, PPN sampling aims to detect, identify, and assess their population levels in soil/plant tissues. The timing, tool, intensity, pattern, and the related material that is sampled all rely on the intended objective and well-conceived scenario to preclude issues of inaccurate/imprecise sampling and the limitation of available funds [20]. Interestingly, heavily PPN-infected plants may lead to an insufficient root system that cannon support numerous PPNs, but samples taken from nearby less infected roots may host more PPNs due to their comparatively large root system. This situation may disrupt the negative correlation usually assumed between crop yield and PPN population levels. Such a disruption might occur when soil samples obtained from the plant rhizosphere are utilized to assess the PPN number per unit (weight in g or volume in cm3). In order to avoid this issue or misleading results, it is advised to count the PPN number per g of fibrous roots in the same soil volume. Otherwise, the discrepancy in the PPN population levels correlated to plant damage may account for a false correlation between the PPN population levels and the growth parameters/productions of infected plants when either weight unit or volume, but not both together, is used. Likewise, using variable samplers for similar integrated pest management (IPM) plans may result in erratic data. Abd-Elgawad [20] elaborated that the area/volume of the sampling units can impact the resulting distribution patterns of the encountered nematodes. Though commonly acceptable, sampling for similar goals is carried out with cores or augers that vary from one trial/location to another. This may also lead to the misinterpretation of data and erratic results. Sampling the same spot with two concentric circles (representing two core diameters) might unintentionally display two different patterns of spatial distribution for the same nematode population (Figure 1). Consequently, counts of the targeted nematode species would require square root (for random distribution) or log (for aggregated distribution) data transformations to stabilize the treatment variances of experimental variables, which is a prerequisite to fulfill the assumptions of parametric statistical techniques, e.g., correlation, regression, and analysis of variance [22]. Moreover, a standardized sampler can provide a reasonable comparison of different experiments and allow the analysis of various individual tests in order to draw substantial and sound conclusions of plant-PPN interactions. To summarize, a 2 cm diameter sampler with adjustable depths was suggested unless the experiment calls for a specific experimental objective [20]. Moreover, the PPN vertical distribution is shaped differently based on the root systems of the host plants. Deep-rooted plants need deeper sampling. For instance, PPNs of grape roots (60 cm root depth) have vertical distribution patterns different from shallow-rooted ones such as squash (20 cm root depth). However, a 30 cm root sampler can generally target the PPNs at their highest and effective densities [20]. Such suggestions avoid other flaws, as the attributes of a distribution pattern mostly rely on the “standard” scale over which it is set. Therefore, relevant suppliers/manufacturers would preferably cooperate with pest control specialists to form/standardize their tools to better configure plant-PPN interactions and IPM schemes. Meanwhile, researchers should use multiple indices of dispersion to supplement each other in order to clarify such interactions and show more aspects of the distribution patterns. Using two indices together, Gorny et al. [23] could set sound sampling schemes and define certain sites to reliably and inexpensively apply nematicide.
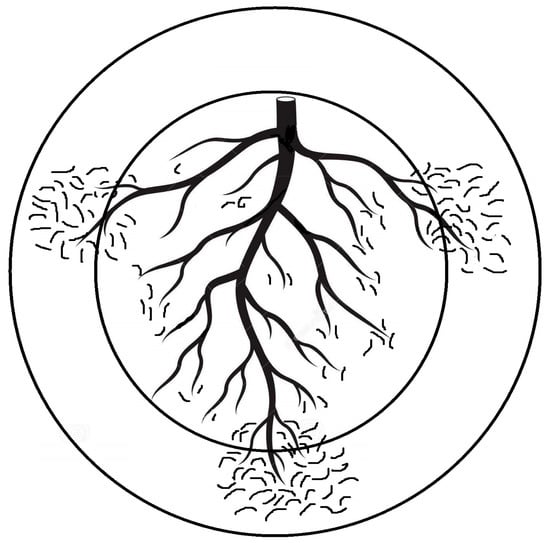
Figure 1. Two quadrat sizes of augers are illustrated by concentric circles. The inner circle displays random nematode distribution around the main plant roots, but the outer circle shows aggregated nematode distribution around lateral fibrous roots [20].
The pros and cons of each PPN extraction method should be considered. Sieving and centrifugation utilizing a sucrose gradient may extract and quantify both dead and live PPNs found in soils. On the contrary, the Baermann funnel technique and its modifications can only separate and extract live nematodes. Clearly, additional tests/bioassays may be needed to find the most reliable extraction technique that is closely related to the examined fauna and flora to study certain plant–nematode interactions [24]. As classical extraction methods inherit imperfect extraction efficiency, a series of extractions via the aforementioned techniques to significantly raise the PPN separation efficiency could reasonably alleviate this issue [20,25].
The sampling and extraction of molecular materials and biochemicals related to PPNs are relatively novel approaches. Relevant assays [25] may extract isozymes or proteins from the PPNs, e.g., for identification, or from their plant hosts, e.g., for determining the enzyme activity of a host genotype/cultivar to explore its incompatible or compatible reactions to PPN infection. These procedures to study plant–nematode interactions may designate susceptible/resistant plant genotypes and detect the role of BCAs in priming the colonized roots against PPNs [26]. Furthermore, sampling techniques to define and assess volatiles in situ in the soil atmosphere can facilitate the exploration of the chemical cues responsible for communication among the different trophic levels of the present organisms. Therefore, they can contribute to not only characterizing plant–nematode interactions but also to conceiving the best IPM program [27]. However, favorable outputs can often be attained with PPNs at a particular developmental stage [20].
2. Exploiting Various Aspects of Plant–Nematode Interactions for PPN Control
2.1. General Aspects of Plant–Nematode Interactions
As the nematodes and their host plants co-evolve in nature, their related genes have presumably balanced co-existence for functioning via the offensive and defensive interactions exerted by the PPNs and the attacked plants, respectively. Although PPN reproduction is possible only if metabolism is shifted to enable nutrient uptake from plant cells, the processes of nematode feeding comprise various morpho-histological, physiological, and biochemical aspects [6]. These processes, as well as structural and molecular depictions of the key events that are involved in the plant-PPN interactions, should be accurately explored for better nematode management. Histopathological, physiological, and biochemical changes have a wide range since the mode of PPN parasitism has presumably evolved from ectoparasite to endoparasite. Thus, the basic disease triangle—that plant harm happens only when a susceptible genotype and its PPNs co-exist in a convenient environment—displays aspects that are focal to numerous techniques to expand the utilization of these different interactions to the favor of the host. Within this broad concept of the interactions, all the aforementioned aspects should be exploited for nematode control as best we can. Clearly, the lesser damage generally caused by ectoparasites compared to endoparasites should not be neglected. Current and emerging approaches to control these ectoparasites should be earnestly tested. Thus, fundamental to the above-mentioned controlling methods, they should be quarantined to block their spread to other areas. Other prophylactic and curing measures to manage these nematodes, in addition to the endoparasitic nematodes, have been generally reviewed [28]. Moreover, specific measures for PPN control have recently been reviewed on important crops such as tomato [29], pepper [30], potato [31], eggplant [32], strawberry [33], and citrus [34].
2.2. Exploiting Plant–Nematode Molecular Interactions for Endoparasitic Nematodes
On the other hand, intensified and sophisticated control measures are being developed for the most damaging endoparasitic sedentary nematodes, and to lesser degree migratory nematodes, as the molecular events involved in their interactions with host plants could be soundly explored e.g., [15,16,17,18,19,28]. The interactions of host transcriptomes with infections of many related PPN species are being massively researched [17,33,34,35,36,37,38], mostly focusing on species of RKNs and CNs [39,40,41,42,43]. The bases of such studies are to improve scientific assumptions for better understanding and prediction of these interactions. However, the current review would rather address how to exploit these interactions for nematode control, focusing on the molecular control of nematodes. This approach does not negate the importance of addressing the fundamental investigations and the related techniques that were developed to harness such interactions for further services in nematode management.
2.2.1. Optimizing Specific Molecular Techniques for Better Nematode Control
In this respect, traditional techniques for the transcriptome analysis of sedentary nematodes are sometimes based on extracting the nematodes from the host tissue before RNA sequencing (RNA-seq) [44]. However, a more useful method is to utilize dual RNA-seq where the roots of the host plants and their invading PPNs are simultaneously sequenced. With the latter method, a PPN effector gene could be discovered, and the transcriptomic datasets between pre-parasitic and parasitic juveniles of Meloidogyne chitwoodi on potato were compared [45]. Thus, the dual RNA-seq could generate a sophisticated analysis of the M. chitwoodi genes displayed during parasitism as well as the encoded foreseen secreted proteins. Additionally, the superior method [45] could considerably decrease the large list of genes in the M. chitwoodi secretome researched by the classical technique [44]. That is because extracting the nematodes from the roots resulted in recording genes that were not linked to parasitism. While it was quite difficult to functionally report ≥ 300 genes via the classical technique, dual RNA-seq was able to efficiently characterize the functions of fewer genes, particularly early in the life stages of the parasite M. chitwoodi [45]. Therefore, it can facilitate more service in PPN management.
Other techniques should also make proper use of the structure/nature of PPN feeding tubes for optimal PPN control. The ultrastructure of these tubes indicates that Meloidogyne spp., but not Heterodera spp., can swallow relatively large transgenic proteins generated by Bacillus thuringiensis [46,47]. As a result, transgenic Cry5B and Cry6A proteins were swallowed by and consequently suppressed M. incognita development and reproduction in fibrous roots of tomato [47,48]. Conversely, such transgenic plant resistance does not operate for H. schachtii infecting plant roots that similarly express the Cry5B protein via B. thuringiensis. That is because the 54 kDa Cry6A protein is too big to be swallowed by the cyst nematodes; the orifice of the H. schachtii feeding tube is narrow: about 23 kDa [49]. Thus, plant pathologists and stakeholders should be aware of such restrictions in using transgenic Cry proteins for managing serious pathogens such as H. schachtii.
Likewise, a variety of diagnostic assays are being developed using the quantitative polymerase chain reaction (qPCR) for the internal transcribed spacer (ITS) of rDNA in numerous nematode species, but relevant flaws should be solved/avoided. For example, the apparent inconsistency of the ITS sequences of the lesion nematodes, Pratylenchus spp., could raise the inevitable risk of obtaining false-negative reactions as variations are found among individuals from the same Pratylenchus species or false-positive reactions are found for fragments from unidentified species [50]. The latter authors emphasized that inaccurate quantification could also happen because sequences of some genes are present in multiple copies of individual nematode cells. Additionally, the copy numbers of particular genes may differ among the developmental stages of the same nematode species [20]. These and other relevant data should be addressed cautiously, as they may participate in obtaining imprecise or misleading molecular PPN–host relationships [39,40,41,42]. Contrariwise, other bioassays and experimental results proved useful but may need further optimization to upgrade safe and effective nematode control. Admittedly, comparative secretome analyses for various PPN species or strains can define molecules that are closely involved in particular aspects of the nematode disease and/or govern PPN virulence in the invaded host. The perfection of these analyses is mandatory to set an optimized molecular control of PPNs [41].
2.2.2. Exploiting RNA Interference for Favorable Plant–Nematode Interactions
Certain genes engaged in the RNA interference (RNAi) pathways of the nematode species could be favorably selected for nematode control [51]. Because RNAi is used to preclude messenger RNA (mRNA) from its role in protein synthesis, it can consequently block the related gene function. Subsequently, silencing several of these functional genes simultaneously in a combinatorial approach could be more impactful in controlling M. incognita on tobacco, Nicotiana tabacum [21]. Moreover, an RNAi technique could be exploited to determine certain nematode effectors to manipulate them for the reliable control of the pinewood nematode Bursaphelenchus xylophilus [52]. The authors used four B. xylophilus isolates that varied in their virulence levels to detect virulence determinants. Four determinants were determined and quantitatively evaluated at the transcript level at different stages, pre-inoculation, 3 days after inoculation (dai), and 7 dai into Pinus thunbergii seedlings, via a real-time reverse-transcription polymerase chain reaction analysis. They found considerably elevated transcript levels of the determinants Bx-CAT1, Bx-CAT2, and Bx-GH30 in virulent B. xylophilus isolates relative to avirulent isolates at two of the three examined stages, that is, at pre-inoculation and 3 dai. Thus, the comparative analyses indicated that Bx-CAT2 and Bx-GH30 share in the B. xylophilus virulence of specific isolates on the susceptible trees. They are significantly involved as nematode effectors in pine wilt disease [52]. Hence, RNAi should be exploited to suppress these effectors, which display themselves as genes responsible for nematode virulence.
Obviously, silencing such effector genes in nematode species that bring about infection and damage of susceptible plants via the RNAi strategy could properly avoid the infection and resulting damage. Following this line of thinking, the RNAi could effectively be used for economically important nematode species [19,21,52,53,54,55,56]. It was applied to silence the Meloidogyne incognita-related effector genes that interact with transcription factors of host plants to operate key enzymes for cell wall degradation. These related M. incognita genes were suppressed, and consequently cell wall degradation was blocked [55]. Clearly, experimentation to exploit plant–nematode interactions via RNAi was proven to upgrade safe and effective nematode control. Therefore, this technique should be wisely manipulated and expanded on a field scale as long as there are no negative consequences on both the environment and the agronomic performance of the transgenic plants. In this respect, Iqbal et al. [51] recommended that the submission of double-stranded RNA (dsRNA) to the intended nematode species via RNA silencing is more workable than any other method (e.g., spraying) for a PPN management strategy. For the perfection of the related processes, they [51] proposed that the original goal, the resistant phenotypes of nematode species/strains, and the current integration advantages of RNAi should further be researched because some of the effector genes may react to RNAi differently. For quite favorable plant–pathogen interactions, it is preferable that an RNAi technique induces broad-spectrum resistance or at least stable resistance to the intended nematode species, i.e., confirmed in the progenies of the subsequent generations of the transgenic plant genotype [56].
2.2.3. Upgrading the Utility of Resistance Genes
The arsenal of PPN resistance (R) genes should be fully exploited and boosted. R genes that were already successfully cloned and transferred from definite plant cultivars to susceptible ones (e.g., Hs1pro−1 from Beta procumbens against H. schachtii, Hero A from Solanum lycopersicum against Globodera rostochiensis, the Mi-1.2 from S. lycopersicum against M. incognita, Gpa-2 from Solanum tuberosum against G. pallida and G. rostochiensis, and Cre loci from Aegilops spp. against Heterodera avenae infecting Triticum aestivum [41,57]) should be further expanded. Other R genes such as Me in pepper, Rk in cowpea, Rhg1 in soybean, Ma in Prunus spp., and Mex1 in coffee have proven benefits in elevating resistance against Meloidogyne spp. [41,58]. Plant resistance to nematodes, as an example to be followed, was acquired in transgenic lettuce, Lactuca sativa, as the Mi-1 gene present in tomato was cloned and transferred to this distant plant species [59]. Clearly, challenges should be faced to extend resistance and its stability and durability via wise the incorporation of proper R genes into economic crops [41]. Because Heterodera glycines is the major soybean pest in intensively and broadly cultivated areas of the world, a supporting action was adopted to compensate for the diminishing plant resistance displayed by a high copy number of the rhg1-b allele. This was materialized via a plant-incorporated protectant, that is, B. thuringiensis delta-endotoxin (Cry14Ab) [60]. As a result, Cry14Ab-expressing soybean plants exhibited fewer H. glycines cyst and egg counts than control plants with only the rhg1-b allele. Such techniques for exploiting plant–nematode interactions for a more favorable support of plant defense could demonstrate the potential of Cry14Ab to control PPNs.
Another challenge is linked to the gene construct itself, e.g., dual or single genes. Genetically engineered tomato plants with PjCHI-1 and CeCPI in double-structured genes and synthetic promoters displayed diminished RKN infection and multiplication compared to their corresponding tomato plants with a single gene [61]. Moreover, the maintenance of durable resistance may be obtained by conveying multiple resistance genes to specific cultivars within IPM programs. Moreover, the recent detection of a nematode-associated molecular pattern (NAMP) plant receptor may be exploited as a useful tool to develop durable plant resistance against economically important PPNs [40,62]. For such programs, safe chemical pesticides, crop rotation, and/or BCAs/their bioactive compounds can aid in decreasing the pressure on resistant plant species/varieties to lessen/avoid the development or emergence of virulent PPN populations.
Likewise, the technique of using an expression quantitative trait locus (eQTL) map to characterize interacting sets of host plant and PPN genes may be harnessed to investigate how the nematode species can fix gene expression differently according to the genetic constitution of the attacked host plant [37]. For better processing, putative targets, defined by Perry and Moens [38], in the PPN life cycles for various life strategies of PPN genera/species should be the focal points for nematode management. Moreover, comparative genomics can increase our knowledge regarding their life cycles, parasitic strategies, and vulnerable life stages. Obviously, further investigation of incompatible PPN-plant interactions should focus on plant species/cultivars with limited resources of available genetic and genomic materials such as commercial full genome arrays, completed genome and transcriptome sequences, and near isogenic and mutant lines [39]. Even with the relatively few detected R genes, there are factors that can break their resistance processes. Among these factors, high temperature, virulent RKN populations, and high PPN population densities were recently reviewed [41]. Therefore, plant breeders and nematologists would preferably rely on activating numerous R genes or QTLs, not a single gene, to circumvent these shortcomings for the economically important plant species [63,64,65].
Eventually, a variety of R genes with various pathways and expressions may cooperate for specific plant phenotypes. Therefore, more knowledge is desperately needed regarding the mechanisms engaged in host-specific resistance/susceptibility formed by PPN effectors and resistance genes or quantitative trait loci to upgrade their reliable and durable use in chief crops. Abd-Elgawad [41] proposed utilizing the effects of these approaches on a case-by-case basis. This will enable biologists to track and improve PPN management based on the existing variables. It would grant us a precise fixing of the factors deciding the gene expressions and functions as well as link them to other PPN control strategies into IPM schemes.
2.2.4. Marker-Assisted Selection to Ease and Perfect Nematode Management
Marker-assisted selection (MAS) is a useful method for both setting the introgression of genes linked to desirable plant traits and favoring the characterization of molecular PPN–host interactions. Therefore, MAS may work in the position of chromosome landmarks and should further be utilized for gene incorporation and stacking. A materialized merit of MAS is its ability to characterize tomato cultivars that possess multiple disease resistance traits. In this example, the Mi-1 homologs can bestow resistance on these cultivars to control a relatively wide range of pests/pathogens. The latter comprise not only the most widespread RKNs (Meloidogyne incognita, M. javanica, and M. arenaria) but also important insects, that is, sweet potato whitefly (Bemisia tabaci), potato aphids (Macrosiphum euphorbiae), and oomycetes (Phytophthora infestans) in tomato plants [66,67]. Thus, a variety of methods, depending on molecular markers for phytonematode resistance, should be developed and effectively utilized to select a wide range of major plant species/varieties for resistance against economically important PPN pests [68,69]. These methods usually include restriction amplified length polymorphisms (RALPs), amplified fragment length polymorphisms (AFLPs), cleaved amplified polymorphic sequences (CAPS), random amplified polymorphic DNA (RAPDs), reverse-transcription polymerase chain reaction (RT-PCR), sequenced characterized amplified regions (SCAR), single-nucleotide polymorphisms (SNPs), simple sequence repeats (SSRs), and sequence-tagged sites (STS). Furthermore, MAS seem to be ready to work, even on a broader genetic pool than the currently limited resistance sources via surveying diverse habitats for novel PPN-R genes. Examples of recent progress in marker-assisted breeding programs could cover not only the main diploid crop species but also polyploid plant species such as sweet potato, Ipomoea batatas. For such a complexity of the polyploid genetic constitution and the highly heterogeneous genome of I. batatas, the genome-wide association technique, which utilizes multiple dose markers to evaluate autopolyploid species was developed [70].
2.2.5. Other Techniques to Facilitate Molecular PPN Control
Emerging and novel techniques should be utilized and developed in favor of plants while interacting with nematodes. For instance, utilizing enzyme inhibitor coding genes such as proteinase inhibitors (PIs) can preclude the operation of proteinases/proteases freed from the nematodes while attacking the plant. Such PIs could be activated against all proteinases, metalloproteinases, aspartic, cysteine, and serine of these nematodes. Techniques to exploit PIs against the nematodes were recently reviewed [81,82]. Other types of anti-nematode proteins, e.g., Bt Cry proteins, several antibodies, and lectins, can be used with various modes of action against PPNs in plants. Therefore, their action to suppress the nematodes should be well-directed, as aforementioned, to optimize PPN control e.g., [47,48,49]. Most of these compounds are generated by BCAs. On the other hand, a few chemodisruptive peptides can disrupt the PPN usage of their chemoreceptive neurons to contact their host plants or move away from their non-hosts. These neurons recognize specific chemical stimuli for attacking and evading their host and non-host plants, respectively. In this respect, acetylcholinesterase (AChE) and/or nicotinic acetylcholine receptors are typically utilized for the proper function of their nervous systems. However, certain peptides, usually at low concentrations, are able to bind with these receptors and subsequently disrupt the nematode’s chemoreception ability via blocking their response to chemical signals [82]. A peptide generated by genetically engineered potato plants could suppress G. pallida-AchE, leading to the disorientation and misguidance of the nematodes when attacking the host plant. The technique resulted in a 52% decrease in the number of G. pallida females [83].
This entry is adapted from the peer-reviewed paper 10.3390/life12111916
This entry is offline, you can click here to edit this entry!
