Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The metagenomic next-generation sequencing (mNGS) method is preferred for genotyping identification of organisms, identification at the species level, illumination of metabolic pathways, and determination of microbiota.
- mNGS
- metabarcoding
- ITS
- bioinformatic analysis
1. Introduction
Farmers worldwide have struggled with crop losses caused by pathogens, including bacteria, viruses, and fungi. The main biotic stress that causes the most economic damage and losses is fungal pathogens. Although the course of the disease and the loss of crops vary according to the host plant, sometimes, up to 100% crop losses are experienced. These losses will pave the way for alleviating food shortages and ecological degradation in the future. Difficulties in culturing and diagnosing organisms are at the forefront of the unavoidable reasons for yield losses. Therefore, it is crucial to have state-of-the-art methods for detecting pathogens and preventing diseases, aiming to reduce crops losses at all stages of crop production (from growth through harvest and postharvest processing) and to ensure agricultural sustainability. Metagenomics is the most direct and unbiased technique to investigate the microbiomes’ functionality, and it is a relatively new addition to the molecular toolkit for pathologists. The term refers to the practice of randomly sequencing the genomic DNA of samples (crop or soil) in an environment, as in the present study [1][2][3]. Subsequently, the development of gene expression techniques that enable the discovery of new genes and metabolic products inspired the “metagenomic” science, which provides all genomic information that can be obtained without culturing under in vitro conditions.
DNA sequencing approaches provide basic information about the diversity of living things of biological importance. Despite their high cost, Sanger sequencing technologies are one of the most preferred methods in sequencing technologies. However, as an alternative to this; many sequencing technologies are widely used, including third or next generation sequencing technologies (NGS) such as Illumina, Ion Torrent, HeliScope, Pacific Biosciences (PacBio), 454/Roche, Sequencing by Oligo Ligation Detection (SOLiD), and Oxford Nanopore. It is preferred, and reduces the high sequencing cost [4]. Next-generation sequencing technologies enable the sequencing of part or all of an organism’s genome. Moreover, metagenomic next-generation sequencing (mNGS) can be used to provide information on the diversity of biologically important resources, analyze DNA sequences, uncover details of metabolic pathways, identify homology-based genes, discover industrially important enzymes, and solve important problems such as the detection of viral and fungal pathogens, among others.
2. Multiple Real-World Applications for mNGS
mNGS technologies can be optimized for use in many areas today (Figure 1). Even if each usage area seems different, mNGS is a common point due to the similarity of the specific barcodes and the method used (Figure 2). One of the primary purposes of mNGS is detecting all culturable and non-culturable substrates and organisms in the medium or host. For this reason, organisms can be scanned via barcodes specific to the species to be determined. There are 16 S rRNA-based universal barcodes used for bacteria, while barcodes from the ITS region are preferred for fungal pathogens. Evolutionary and ecological studies have a vital role in the development of metagenomic science. The first discovery of proteorhodopsin proteins occurred in environmental DNA. Complete genome data of microbial communities found in environmental samples can be obtained today, with scientists aiming to reveal whole genomes.
Figure 1. Applications of mNGS technology in different fields.
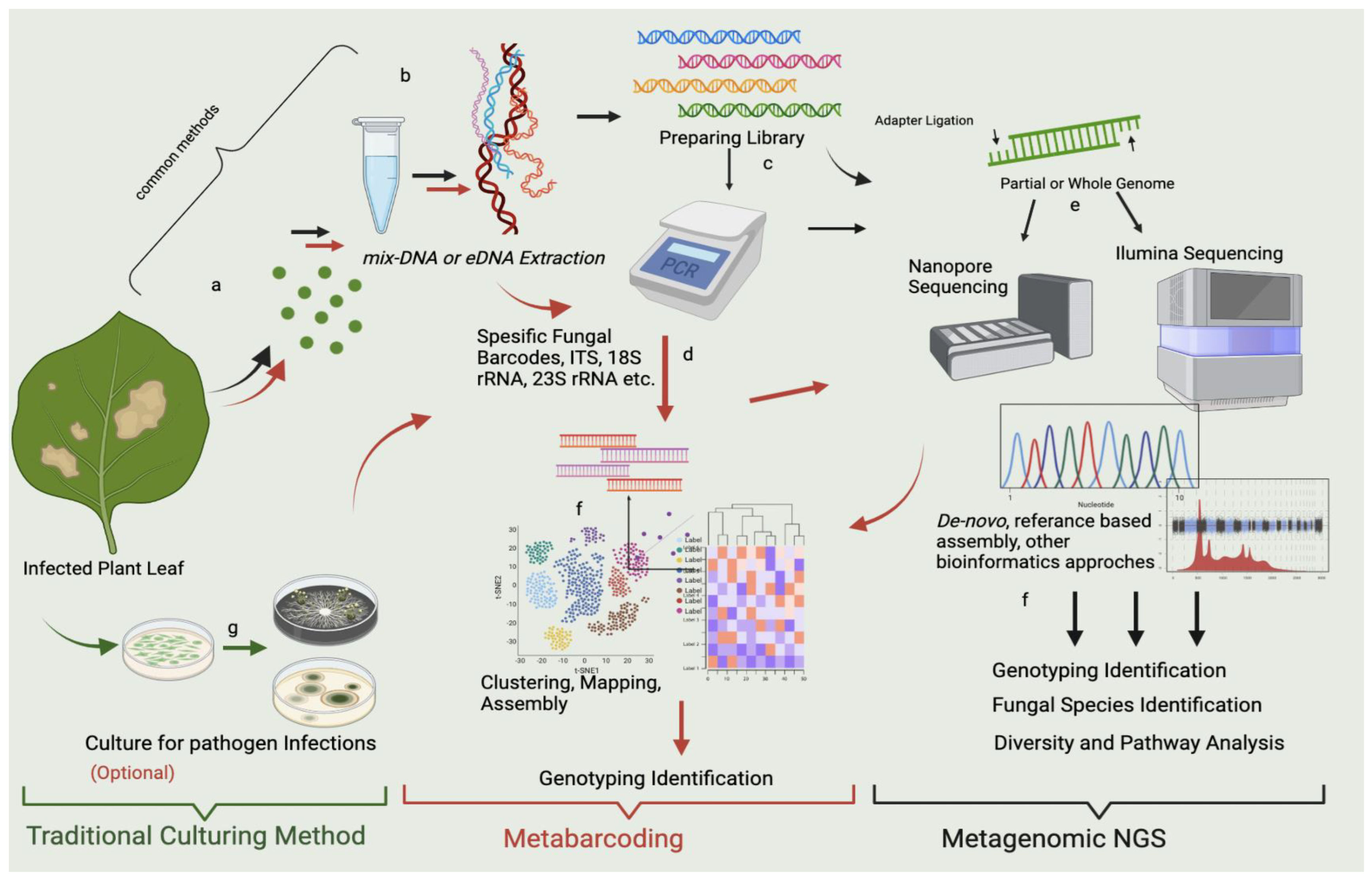
Figure 2. mNGS and metabarcoding workflow chart for the sample obtained from the infected leaf. The workflow highlighted in red shows metabarcoding pathways, which use specific metabarcodes for fungal detection, and the “black” arrow shows the mNGS pathways. In the workflows, the Polymerase Chain Reaction (PCR) stage is optional, and after sequencing and bioinformatic analysis, metabarcoding shows genotyping identification. mNGS indicates fungal species identification, microbial diversity, pathway detection, and genotyping identification. Both techniques seem to include the same steps; however, the algorithms (the bioinformatics analysis) differ. In metabarcoding, certain parts of the genome are sequenced using target-specific barcodes, and in mNGS, either a partial or a whole genome is sequenced by reference-based comparison with the prepared library. Both approaches provide a fundamental approach and solution for metagenomics. The workflow highlighted in green represents the traditional culturing method at the researcher’s discretion. It may allow culturing of some of the possible microorganisms prior to mNGS and metabarcoding. However, this gives an assignment far below sufficient for mNGS and metabarcoding. The stages represented in the figure can be summarized as follows: (a) sampling of infected parts of the plant (leaf discs are preferred); (b) DNA extraction from leaf; (c) library preparation; (d) PCR amplification of gene regions of microbial pathogens with specific gene barcodes; (e) sequencing with Illumina, Nanopore, etc.; (f) bioinformatic analysis of mNGS containing de novo approaches and referenced based assembly, bioinformatic analysis for metabarcoding assembly, clustering, and prediction; and (g) control culture of infected leaves.
According to the review of the literature, metabarcoding or metagenomic sciences have been widely used in health sciences up to now [5]. The first use of metagenomics in health sciences dates back to 2008 [6]. After the organ transplant of three different patients, the accompanying analysis showed Arenavirus in recipients using the mNGS method. Following this report, mNGS became a routinely accepted method for detecting infectious diseases to date [7]. By analyzing body fluids [8], detecting pulmonary infection in lung tissues [9][10] and microbial organisms underlying chronic meningitis, determining organisms causing tuberculous meningitis in cerebrospinal fluid [11], and even identifying pathogens responsible for uncultured prosthetic joint infection [12] have become trends of choice. Most of the studies use viral, bacterial, and fungal kits. In addition to the detection of different infections, scientists aimed to map human-associated microbial communities, such as the gut, mouth, skin, and vagina, as part of the Human Microbiome project [13][14]. mNGS technology is also used in forensic sciences, particularly in the resolution of forensic cases such as geographic locations and surface analysis [15][16], identification [17][18], biological sex determination [19][20], trace evidence [21], manner and cause of death determination [22][23], and postmortem microbiota determination are becoming more and more common [24][25].
mNGS technologies in the agricultural and industrial fields have led to important discoveries. New generation sequencing studies, primarily available in plant roots, are increasingly preferred, as they enable the discovery of important secondary metabolites, enzymes, and metabolites [26]. With the influence of industrial applications of the metagenomic approach, the discovery of stress-sensitive bioactive compounds reveals the genetic information of organisms living in extreme conditions. This discovery is used for efficient crop production and elucidation of plant stress mechanisms.
Agronomically, the scope of mNGS technologies is expanding day by day. In a previous report, microbial diversity data are essential for sustainable black pepper production [27]. The organisms that make up the plant microbiota provide the necessary nutrients for the growth and development of the plant. Therefore, mNGS technologies are vital for the sustainability of agriculture. Moreover, using metagenomic data to detect and control biotic stress factors affecting crop yield offers optimistic promises for the future. For example, the metagenomic method with 16 S rRNA barcodes was applied to samples obtained from black pepper roots grown in Vietnam [27], with promising outcomes.
This entry is adapted from the peer-reviewed paper 10.3390/jof8111195
References
- Woese, C.R. The problem of evolving a genetic code. Bioscience 1970, 20, 471–485.
- Walters, W.A.; Knight, R. Technology and techniques for microbial ecology via DNA sequencing. Ann. Am. Thorac. Soc. 2014, 11 (Suppl. S1), S16–S20.
- Yadav, S.; Gettu, N.; Swain, B.; Kumari, K.; Ojha, N.; Gunthe, S.S. Bioaerosol impact on crop health over India due to emerging fungal diseases (EFDs): An important missing link. Environ. Sci. Pollut. Res. 2020, 27, 12802–12829.
- Ller, R.R.; Montoya, V.; Gardy, J.L.; Patrick, D.M.; Tang, P. Metagenomics for pathogen detection in public health. Genome Med. 2013, 5, 1–14.
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 2019, 14, 319–338.
- Palacios, G.; Druce, J.; Du, L.; Tran, T.; Birch, C.; Briese, T.; Lipkin, W.I. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 2008, 358, 991–998.
- Han, D.; Li, Z.; Li, R.; Tan, P.; Zhang, R.; Li, J. mNGS in clinical microbiology laboratories: On the road to maturity. Crit. Rev. Microbiol. 2019, 45, 668–685.
- Gu, W.; Deng, X.; Lee, M.; Sucu, Y.D.; Arevalo, S.; Stryke, D.; Federman, S.; Gopez, A.; Reyes, K.; Zorn, K.; et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 2021, 27, 115–124.
- Li, H.; Gao, H.; Meng, H.; Wang, Q.; Li, S.; Chen, H.; Li, Y.; Wang, H. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front. Cell. Infect. Microbiol. 2018, 8, 205.
- Qian, Y.-Y.; Wang, H.-Y.; Zhou, Y.; Zhang, H.-C.; Zhu, Y.-M.; Zhou, X.; Ying, Y.; Cui, P.; Wu, H.-L.; Zhang, W.-H.; et al. Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front. Cell. Infect. Microbiol. 2021, 10, 567615.
- Wang, S.; Chen, Y.; Wang, D.; Wu, Y.; Zhao, D.; Zhang, J.; Xie, H.; Gong, Y.; Sun, R.; Nie, X.; et al. The feasibility of metagenomic next-generation sequencing to identify pathogens causing tuberculous meningitis in cerebrospinal fluid. Front. Microbiol. 2019, 10, 1993.
- Wang, C.; Huang, Z.; Li, W.; Fang, X.; Zhang, W. Can metagenomic next-generation sequencing identify the pathogens responsible for culture-negative prosthetic joint infection? BMC Infect. Dis. 2020, 20, 1–7.
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome projects. Nature 2007, 449, 804–810.
- Gevers, D.; Knight, R.; Petrosino, J.F.; Huang, K.; McGuire, A.L.; Birren, B.W.; Nelson, K.E.; White, O.; Methé, B.A.; Huttenhower, C. The Human Microbiome Project: A community resource for the healthy human microbiome. PLoS Biol. 2012, 10, e1001377.
- Habtom, H.; Pasternak, Z.; Matan, O.; Azulay, C.; Gafny, R.; Jurkevitch, E. Applying microbial biogeography in soil forensics. Forensic Sci. Int. Genet. 2019, 38, 195–203.
- Keet, J.H.; Ellis, A.G.; Hui, C.; Le Roux, J.J. Strong spatial and temporal turnover of soil bacterial communities in South Africa’s hyperdiverse fynbos biome. Soil Biol. Biochem. 2019, 136, 107541.
- Wang, S.; Song, F.; Wang, Y.; Huang, Y.; Xie, B.; Luo, H. High resolution melting analysis (HRM) based on 16SrRNA as a tool for personal identification with the human oral microbiome. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 161–163.
- Schmedes, S.E.; Woerner, A.E.; Novroski, N.M.; Wendt, F.R.; King, J.L.; Stephens, K.M.; Budowle, B. Targeted sequencing of clade-specific markers from skin microbiomes for forensic human identification. Forensic Sci. Int. Genet. 2018, 32, 50–61.
- Bell, C.R.; Wilkinson, J.E.; Robertson, B.K.; Javan, G.T. Sex-related differences in the thanatomicrobiome in postmortem heart samples using bacterial gene regions V1-2 and V4. Lett. Appl. Microbiol. 2018, 67, 144–153.
- Zhou, W.; Bian, Y. Thanatomicrobiome composition profiling as a tool for forensic investigation. Forensic Sci. Res. 2018, 3, 105–110.
- Robinson, J.M.; Pasternak, Z.; Mason, C.E.; Elhaik, E. Forensic applications of microbiomics: A review. Front. Microbiol. 2021, 11, 3455.
- Zhang, Y.; Pechal, J.L.; Schmidt, C.J.; Jordan, H.R.; Wang, W.W.; Benbow, M.E.; Sze, S.-H.; Tarone, A. Machine learning performance in a microbial molecular autopsy context: A cross-sectional postmortem human population study. PLoS ONE 2019, 14, e0213829.
- Marella, G.L.; Feola, A.; Marsella, L.T.; Mauriello, S.; Giugliano, P.; Arcudi, G. Diagnosis of drowning, an everlasting challenge in forensic medicine: Review of the literature and proposal of a diagnostic algorithm. Acta Med. Int. 2019, 35, 900–919.
- Metcalf, J.L.; Parfrey, L.W.; Gonzalez, A.; Lauber, C.L.; Knights, D.; Ackermann, G.; Humphrey, G.C.; Gebert, M.J.; Van Treuren, W.; Berg-Lyons, D.; et al. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. Elife 2013, 2, e01104.
- Pechal, J.L.; Crippen, T.L.; Benbow, M.E.; Tarone, A.M.; Dowd, S.; Tomberlin, J.K. The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int. J. Leg. Med. 2014, 128, 193–205.
- Lorenz, P.; Eck, J. Metagenomics and industrial applications. Nat. Rev. Microbiol. 2005, 3, 510–516.
- Tran, D.M.; Nguyen, T.H.; Huynh, T.U.; Do, T.O.; Nguyen, Q.V.; Nguyen, A.D. Analysis of endophytic microbiome dataset from roots of black pepper (Piper nigrum L.) cultivated in the Central Highlands region, Vietnam using 16S rRNA gene metagenomic next-generation sequencing. Data Brief 2022, 42, 108108.
This entry is offline, you can click here to edit this entry!
