Skin is the largest organ of the human body and is a great shield, as it protects it from external infections (environmental and chemical pollutants) as well as from UV irradiation. However, it is vulnerable since its degradation can occur both due to extrinsic and intrinsic factors, leading to early aging. Among all, extrinsic skin aging, called photoaging, is a remarkable result of oxidative stress caused by UV irradiation. In addition, reactive oxygen species (ROS) have also been found to contribute to skin aging, as they are produced in skin cells through UV irradiation, although at low concentrations they could be beneficial for some signaling pathways. Environmental and chemical pollutants also produce ROS, triggering a number of pathologies. Skin’s connective tissue includes a number of constituents, including collagen fibrils, elastic fibers, glycoproteins, and glycosaminoglycans. Among all, proteins like elastin, collagen, the glycosaminoglycan hyaluronic acid, and a polymeric pigment called melanin play pivotal roles in the regulation of skin’s elasticity as well as its protection against UV irradiation.
- database
- natural products
- medicinal plants
- antioxidants
- natural inhibitors
- anti-agings
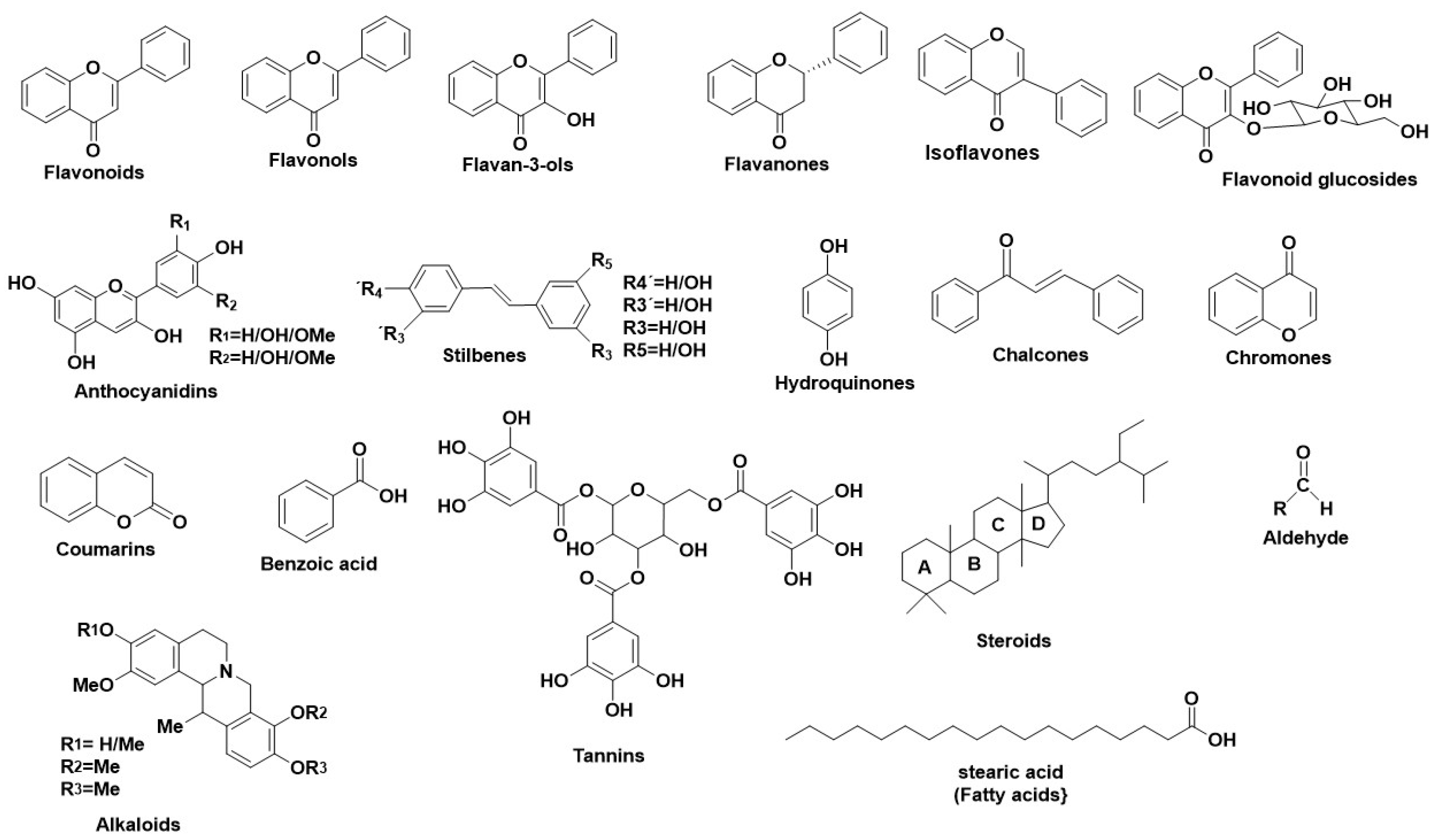
1. Human Neutrophil Elastase (HNE)-A Serine Protease
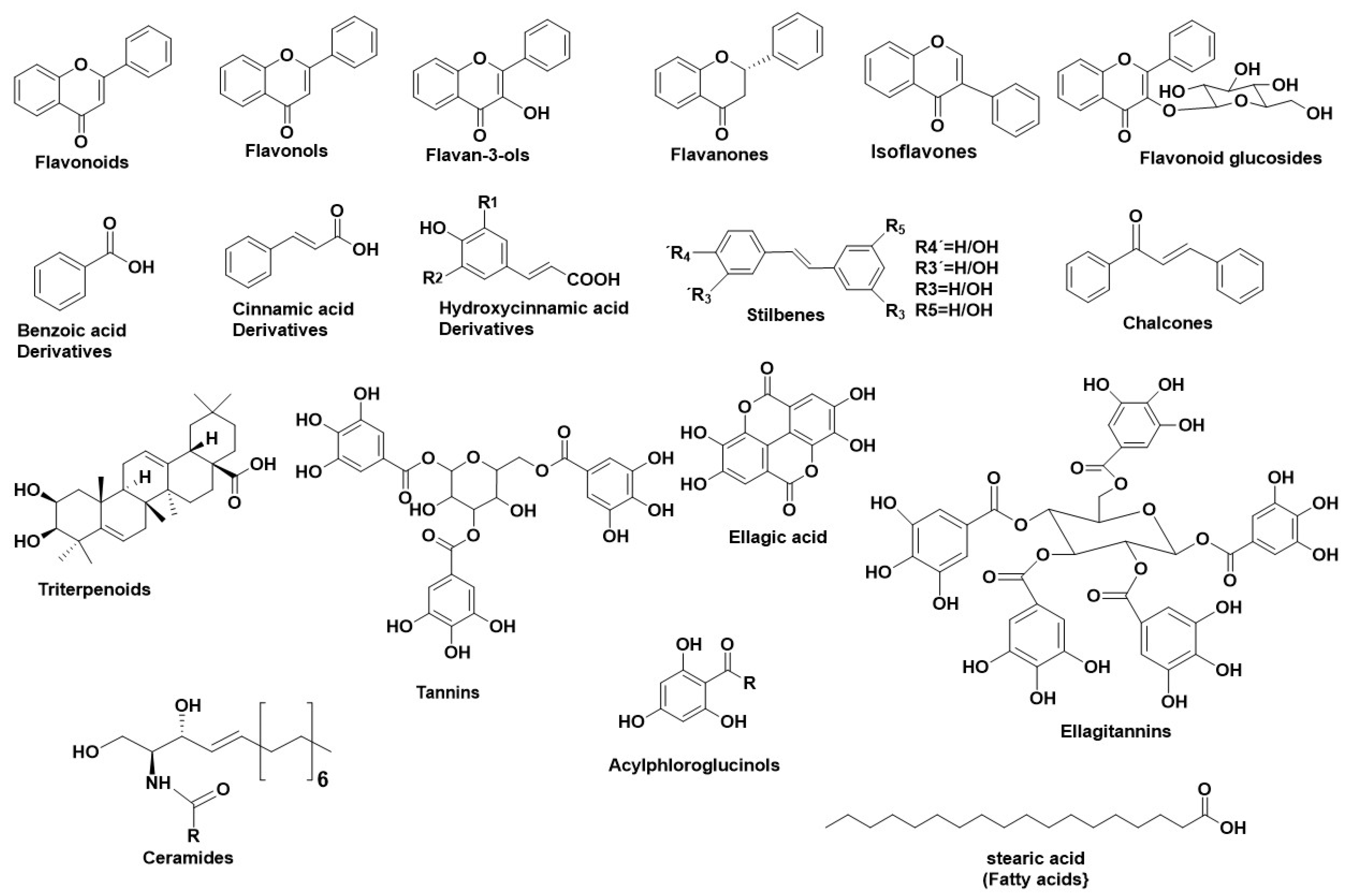
2. Hyaluronidase (Hyal)—A Glycosyl Hydrolase
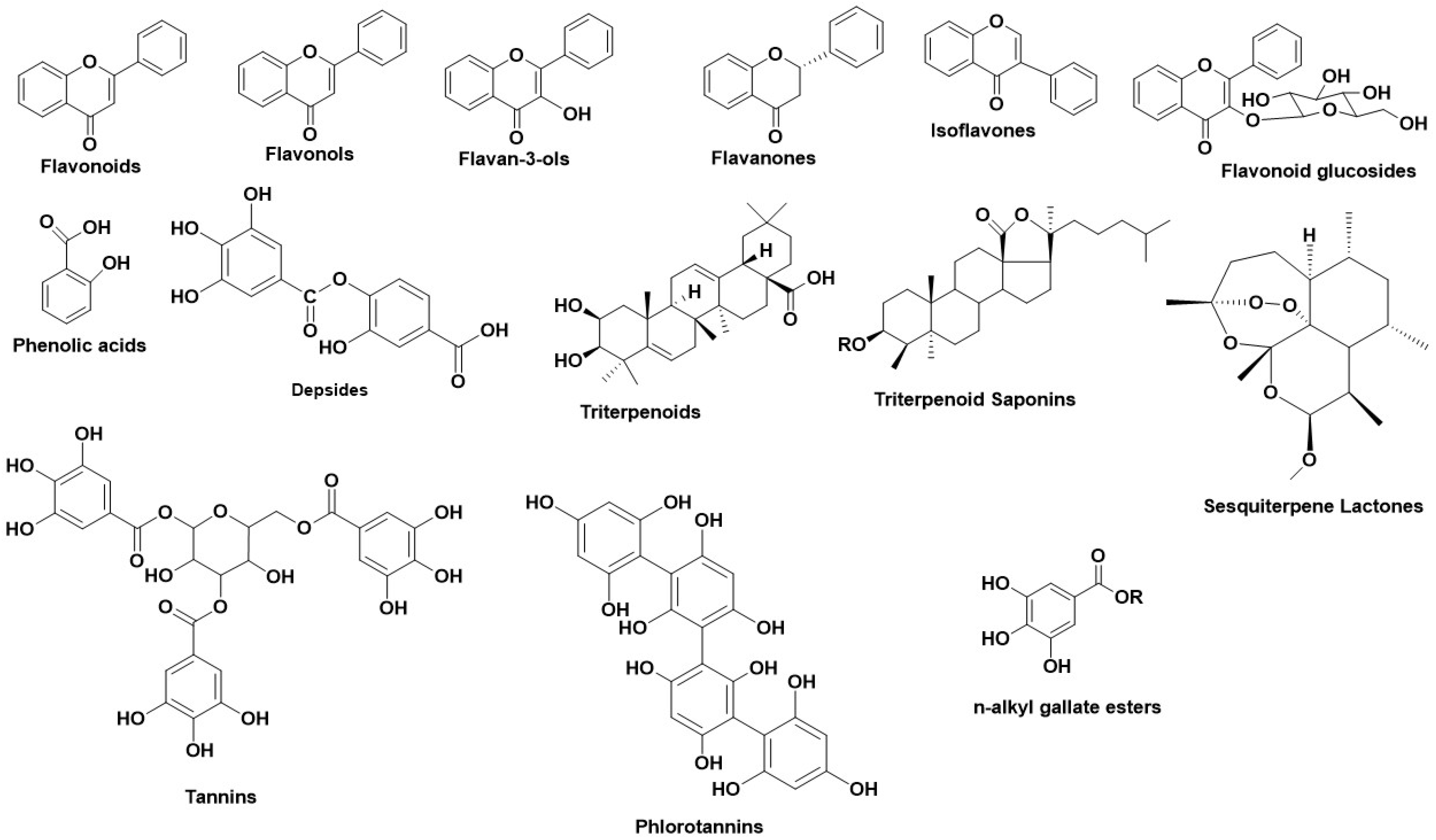
3. Tyrosinase—A Polyphenol Oxidase
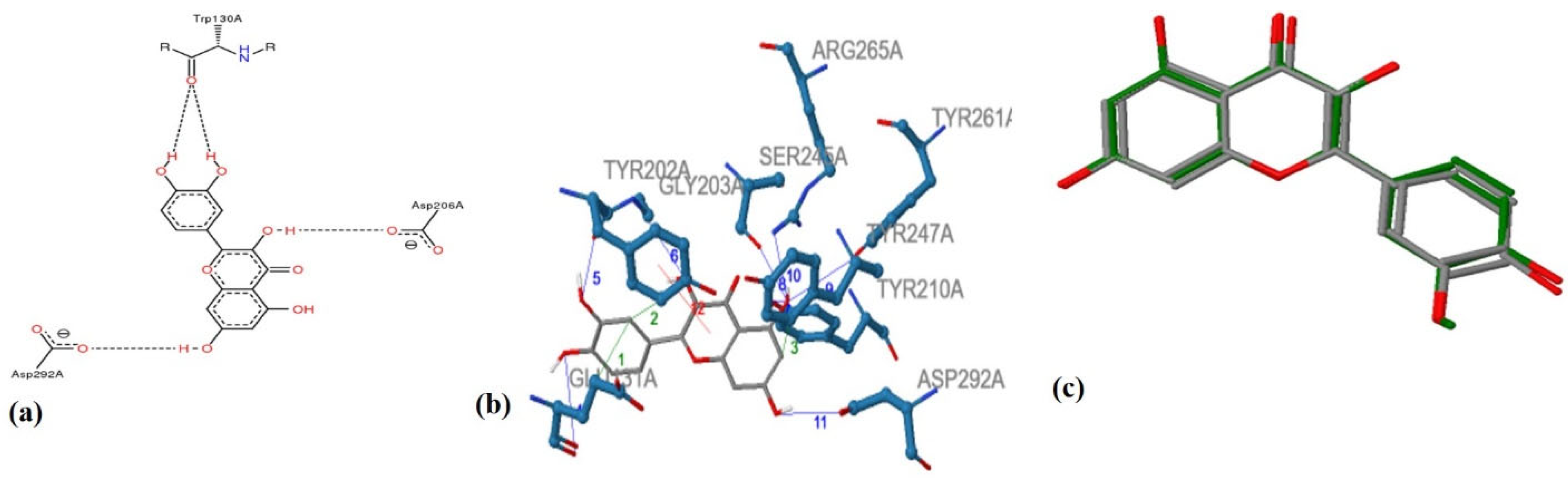
This entry is adapted from the peer-reviewed paper 10.3390/antiox11112268
References
- Bieth, J.G. The elastases. J. Soc. Biol. 2001, 195, 173–179. PMID: 11723830
- Takahashi, H.; Nukiwa, T.; Yoshimura, K.; Quick, C.D.; States, D.J.; Holmes, M.D.; Whang-Peng, J.; Knutsen, T.; Crystal, R.G. Structure of the human neutrophil elastase gene. J. Biol. Chem. 1988, 263, 14739–14747. PMID: 2902087
- Melzig, M.F.; Löser, B.; Ciesielski, S. Inhibition of neutrophil elastase activity by phenolic compounds from plants. Pharmazie 2001, 56, 967–970. PMID: 11802662
- Siedle, B.; Cisielski, S.; Murillo, R.; Lo, B.; Castro, V.; Klaas, C.A.; Hucke, O.; Labahn, A.; Melzig, M.F.; Merfort, I.; et al. Sesquiterpene Lactones as Inhibitors of Human Neutrophil Elastase. Bioorg. Med. Chem. 2002, 10, 2855–2861. DOI: 10.1016/s0968-0896(02)00149-9
- Kim, Y.-J.; Uyama, H.; Kobayashi, S. Inhibition effects of (+)-catechin-aldehyde polycondensates on proteinases causing proteolytic degradation of extracellular matrix. Biochem. Biophys. Res. Commun. 2004, 320, 256–261. DOI: 10.1016/j.bbrc.2004.05.163
- Thring, T.S.; Hili, P.; Naughton, D.P. Anti-collagenase, anti-elastase and anti-oxidant activities of extracts from 21 plants. BMC Complement. Altern. Med. 2009, 9, 27. DOI: 10.1186/1472-6882-9-27
- Belaaouaj, A.; Kim, K.S.; Shapiro, S.D. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science 2000, 289, 1185–1188. DOI: 10.1126/science.289.5482.1185
- Ying, Q.L.; Rinehart, A.R.; Simon, S.R.; Cheronis, J.C. Inhibition of human leucocyte elastase by ursolic acid. Evidence for a binding site for pentacyclic triterpenes. Biochem. J. 1991, 277, 521–526. DOI: 10.1042/bj2770521
- Bode, W.; Edgar Meyer, J.C.P., Jr. Perspectives in Biochemistry. Human Leukocyte and PorcinePancreatic Elastase: X-ray Crystal Structures, Mechanism, Substrate Specificity. Biochemistry 1989, 28, 1951–1963. DOI: 10.1021/bi00431a001
- Feng, L.; Liu, X.; Zhu, W.; Guo, F.; Wu, Y.; Wang, R.; Chen, K.; Huang, C.; Li, Y. Inhibition of human neutrophil elastase by pentacyclic triterpenes. PLoS ONE 2013, 8, e82794. DOI: 10.1371/journal.pone.0082794
- Tamada, T.; Kinoshita, T.; Kurihara, K.; Adachi, M.; Ohhara, T.; Imai, K.; Kuroki, R.; Tada, T. Combined high-resolution neutron and X-ray analysis of inhibited elastase confirms the active-site oxyanion hole but rules against a low-barrier hydrogen bond. J. Am. Chem. Soc. 2009, 131, 11033–11040. DOI: 10.1021/ja9028846
- Hess, G.P.; McConn, J.; Ku, E.; McConkey, G. Studies of the activity of chymotrypsin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1970, 257, 89–104. DOI: 10.1098/rstb.1970.0011
- Siedle, B.; Hrenn, A.; Merfort, I. Natural compounds as inhibitors of human neutrophil elastase. Planta Med. 2007, 73, 401–420. DOI: 10.1055/s-2007-967183
- Löser, B.; Kruse, S.O.; Melzig, M.F.; Nahrstedt, A. Inhibition of neutrophil elastase activity by cinnamic acid derivatives from Cimicifuga racemosa. Planta Med. 2000, 66, 751–753. DOI: 10.1055/s-2000-9563
- Melzig, M.F.; Löser, B.; Lobitz, G.O.; Tamayo-Castillo, G.; Merfort, I. Inhibition of granulocyte elastase activity by caffeic acid derivatives. Pharmazie 1999, 54, 712. PMID: 10522278
- Hamburger, M.; Riese, U.; Graf, H.; Melzig, M.F.; Ciesielski, S.; Baumann, D.; Dittmann, K.; Wegner, C. Constituents in Evening Primrose Oil with Radical Scavenging, Cyclooxygenase, and Neutrophil Elastase Inhibitory Activities. J. Agric. Food Chem. 2002, 50, 5533–5538. DOI: 10.1021/jf025581l
- Xing, X.; Yang, X.; Cao, Y. Study of Ellagic Acid as a Natural Elastase Inhibitor by Spectroscopic Methods. J. Appl. Spectrosc. 2016, 83, 149–155. DOI:https://doi.org/10.1007/s10812-016-0259-4
- Rennert, B.; Melzig, M.F. Free fatty acids inhibit the activity of Clostridium histolyticum collagenase and human neutrophil elastase. Planta Med. 2002, 68, 767–769. DOI: 10.1055/s-2002-34411
- Bizot-Foulon Godeau, G.; Guessous, F.; Lati, E.; Rousset, G.; Roch-Arveillier, M.; Hornebeck, W.V. Inhibition of human neutrophil elastase by wheat ceramides. Int. J. Cosmet. Sci. 1995, 17, 255–264. DOI: 10.1111/j.1467-2494.1995.tb00130.x
- Sim, G.S.; Lee, B.C.; Cho, H.S.; Lee, J.W.; Kim, J.H.; Lee, D.H.; Kim, J.-H.; Pyo, H.B.; Moon, D.C.; Oh, K.W.; et al. Structure activity relationship of antioxidative property of flavonoids and inhibitory effect on matrix metalloproteinase activity in UVA-irradiated human dermal fibroblast. Arch. Pharmacol. Res. 2007, 30, 290–298. DOI: 10.1007/BF02977608
- Wittenauer, J.; Mäckle, S.; Sußmann, D.; Schweiggert-Weisz, U.; Carle, R. Inhibitory effects of polyphenols from grape pomace extract on collagenase and elastase activity. Fitoterapia 2015, 101, 179–187. DOI: 10.1016/j.fitote.2015.01.005
- Załuski, D.; Cieśla, Ł.; Janeczko, Z. Chapter 7—The Structure—Activity Relationships of Plant Secondary Metabolites with Antimicrobial, Free Radical Scavenging and Inhibitory Activity toward Selected Enzymes. Stud. Nat. Prod. Chem. 2015, 45, 217–249. DOI:10.1016/B978-0-444-63473-3.00007-1
- Marković-Housley, Z.; Miglierini, G.; Soldatova, L.; Rizkallah, P.J.; Müller, U.; Schirmer, T. Crystal structure of hyaluronidase, a major allergen of bee venom. Structure 2000, 8, 1025–1035. DOI: 10.1016/s0969-2126(00)00511-6
- Laurent, T.C.; Fraser, J.R. Hyaluronan. FASEB J. 1992, 6, 2397–2404. PMID: 1563592
- Meyer, K. Hyaluronidases. In The Enzymes; Academic Press: New York, NY, USA, 1971; pp. 307–320. DOI: 10.1021/cr050247k
- Stern, R. Devising a pathway for hyaluronan catabolism: Are we there yet? Glycobiology 2003, 13, 105–115. DOI: 10.1093/glycob/cwg112
- Orlando, Z.; Lengers, I.; Melzig, M.F.; Buschauer, A.; Hensel, A.; Jose, J. Autodisplay of human hyaluronidase Hyal-1 on Escherichia coli and identification of plant-derived enzyme inhibitors. Molecules 2015, 20, 15449–15468. DOI: 10.3390/molecules200915449
- Jedrzejas, M.J.; Stern, R. Structures of vertebrate hyaluronidases and their unique enzymatic mechanism of hydrolysis. Proteins 2005, 61, 227–238. DOI: 10.1002/prot.20592
- Csoka, A.B.; Frost, G.I.; Stern, R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001, 20, 499–508. DOI: 10.1016/s0945-053x(01)00172-x
- Chao, K.L.; Muthukumar, L.; Herzberg, O. Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme involved in tumor growth and angiogenesis. Biochemistry 2007, 46, 6911–6920. DOI: 10.1021/bi700382g
- Lokeshwar, V.B.; Rubinowicz, D.; Schroeder, G.L.; Forgacs, E.; Minna, J.D.; Block, N.L.; Nadji, M.; Lokeshwar, B.L. Stromal and epithelial expression of tumor markers hyaluronic acid and HYAL1 hyaluronidase in prostate cancer. J. Biol. Chem. 2001, 276, 11922–11932. DOI: 10.1074/jbc.M008432200
- Lokeshwar, V.B.; Obek, C.; Pham, H.T.; Wei, D.; Young, M.J.; Duncan, R.C.; Soloway, M.S.; Block, N.L. Urinary hyaluronic acid and hyaluronidase: Markers for bladder cancer detection and evaluation of grade. J. Urol. 2000, 163, 348–356. DOI: 10.1016/s0022-5347(05)68050-0
- Tan, J.X.; Wang, X.Y.; Li, H.Y.; Su, X.L.; Wang, L.; Ran, L.; Zheng, K.; Ren, G.S. HYAL1 overexpression is correlated with the malignant behavior of human breast cancer. Int. J. Cancer 2011, 128, 1303–1315. DOI: 10.1002/ijc.25460
- Mio, K.; Stern, R. Inhibitors of the hyaluronidases. Matrix Biol. 2002, 21, 31–37. DOI: 10.1016/s0945-053x(01)00185-8
- Jentsch, H.; Pomowski, R.; Kundt, G.; Göcke, R. Treatment of gingivitis with hyaluronan. J. Clin. Periodontol. 2003, 30, 159–164. DOI: 10.1034/j.1600-051x.2003.300203.x
- Stern, R.; Jedrzejas, M.J. Hyaluronidases: Their genomics, structures, and mechanisms of action. Chem. Rev. 2006, 106, 818–839. DOI: 10.1021/cr050247k
- Hunnicutt, G.R.; Primakoff, P.; Myles, D.G. Sperm surface protein PH-20 is bifunctional: One activity is a hyaluronidase and a second, distinct activity is required in secondary sperm-zona binding. Biol. Reprod. 1996, 55, 80–86. DOI: 10.1095/biolreprod55.1.80
- Lee, S.Y.; Baek, N.; Nam, T.G. Natural, semisynthetic and synthetic tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2016, 31, 1–13. DOI: 10.3109/14756366.2015.1004058
- Jackman, M.P.; Hajnal, A.; Lerch, K. Albino mutants of Streptomyces glaucescens tyrosinase. Biochem. J. 1991, 274 Pt 3, 707–713. DOI: 10.1042/bj2740707
- van Gelder, C.W.; Flurkey, W.H.; Wichers, H.J. Sequence and structural features of plant and fungal tyrosinases. Phytochemistry 1997, 45, 1309–1323. DOI: 10.1016/s0031-9422(97)00186-6
- Himmelwright, R.S.; Eickman, N.C.; Lu Bien, C.D.; Lerch, K.; Solomon, E. Chemical and Spectroscopic Studies of the Binuclear Copper Active Site of Neurospora Tyrosinase: Comparison to Hemocyanins. J. Am. Chem. Soc. 1980, 102, 7339–7344. DOI: https://doi.org/10.1021/ja00544a031
- Magnus, K.A.; Hazes, B.; Ton-That, H.; Bonaventura, C.; Bonaventura, J.; Hol, W.G.J. Crystallographic Analysis of Oxygenated and Deoxygenated States of Arthropod Hemocyanin Shows Unusual Differences. Genet. Proteins Struct. Funct. 1994, 19, 302–309. DOI: 10.1002/prot.340190405
- Volbeda, A.; Hol, W.G. Crystal Structure of Hexameric Haemocyanin from Panulirus Interruptus Refined at 3.2 A Resolution. J. Mol. Biol. 1989, 209, 249–279. DOI: 10.1016/0022-2836(89)90276-3
- Wang, C.; Yan, S.; Huang, R.; Feng, S.; Fu, B.; Weng, X.; Zhou, X. A turn-on fluorescent probe for detection of tyrosinase activity. Analyst 2013, 138, 2825–2828. DOI: 10.1039/c3an00272a
