Cancer cells typically have elevated proteotoxic stress as a result of genomic instability. The disruption of protein homeostasis causes endoplasmic reticulum stress. If not promptly managed, it could lead to a global decline in cellular function and eventual cell death[1]. This summary of review explores different protein quality control pathways and the translation of drugs targeting proteotoxic stress in haematologic cancers (using multiple myeloma as an example) versus solid cancers (using Triple Negative Breast Cancer as an example).
- proteotoxic stress
- protein quality control
- proteasome
- proteasome inhibition
- autophagy
- aggresome
- unfolded protein response
- multiple myeloma
- triple negative breast cancer
1. Proteotoxic stress and cancer
Cancer cells typically have elevated proteotoxic stress as a result of genomic instability. This property has led to the discovery and development of new anticancer therapies. The disruption of protein homeostasis causes endoplasmic reticulum (ER) stress. If not promptly managed, it could lead to a global decline in cellular function and eventual cell death[1].
The protein synthesis process is extremely prone to errors. About 30% of newly synthesised proteins are degraded by the proteasome within minutes of protein translation in mammalian cells[2]. These rapidly degraded proteins are called defective ribosomal proteins (DRiPs) or rapidly degraded polypeptides (RDPs) [3]. In comparison to normal cells, cancer cells generally synthesise proteins (and therefore DRiPs or RDPs) more rapidly due to increased cell division coupled to cell growth [4]. If DRiPs or RDPs are not removed, the basal proteasome load and cellular stress can increase considerably.
2. Protein quality control pathways
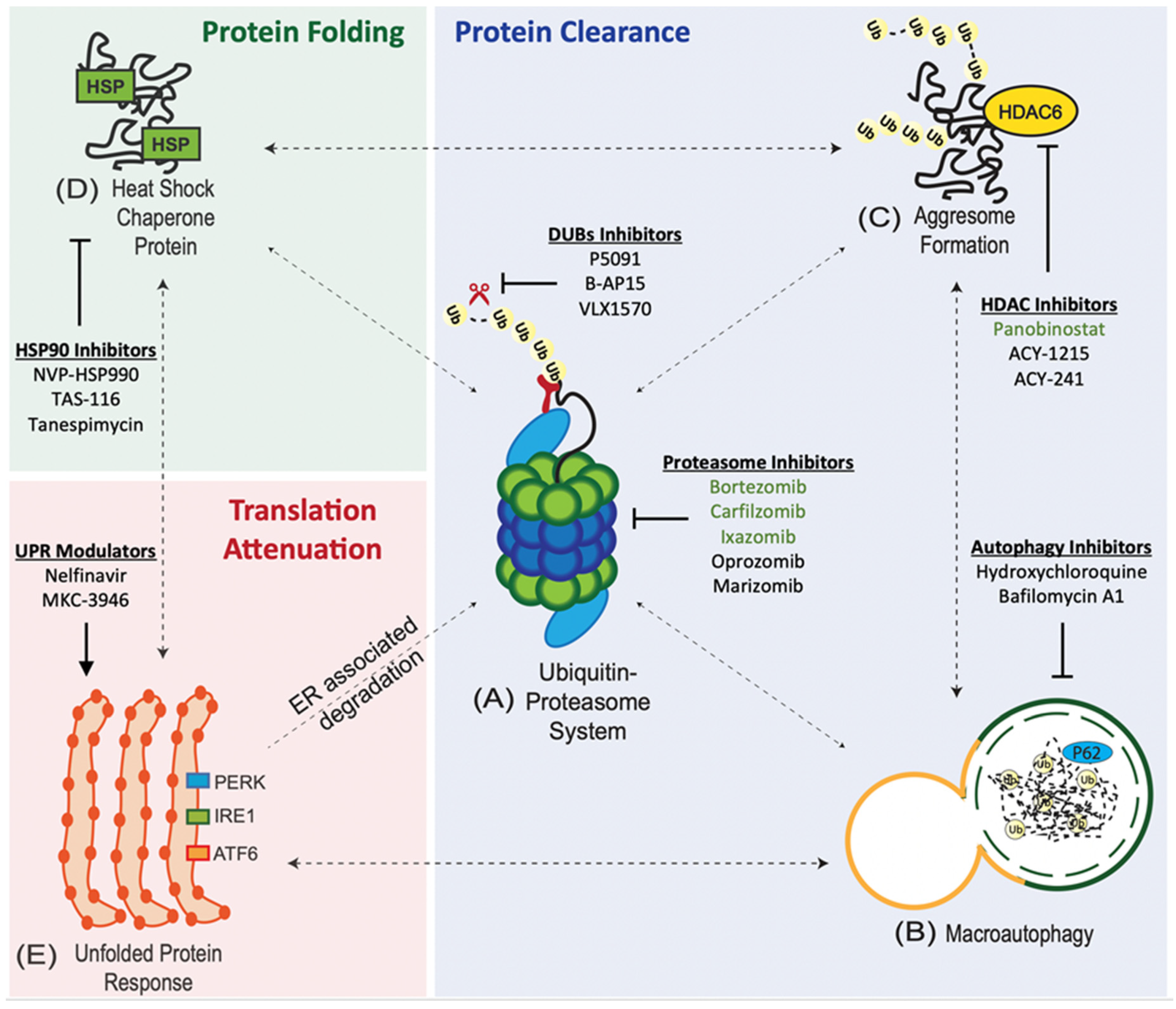
Protein quality control pathways play an important role in tumours due to the intrinsic genetic instability and hostile tumour microenvironment [5]. Cancer cells maintain the integrity of the proteome through an interconnected network of proteostasis pathways.
1. Ubiquitin Proteasome System (UPS). UPS is at the core of proteostasis pathways, and is involved in the degradation of both normal and misfolded proteins [5]. The UPS involves a 3-enzyme-cascade: E1 (activating), E2 (conjugating), and E3 (ligating) enzymes [6]. The targeted (misfolded) proteins are polyubiquitinated or tagged by these 3 enzymes and subsequently degraded by the 26S proteasome [7].
2. Aggresome Pathway. Polyubiquitinated (misfolded) proteins that coalesce to form aggresomal particles are transported towards the microtubule organising center (MTOC) and undergo degradation by autophagy or refolding by the heat-shock protein (HSP) chaperone system [8].
3. Macroautophagy (Autophagy-Lysosome System). Misfolded proteins are taken up by autophagosomes and degraded by lysosomal hydrolases upon fusion with lysosomes [9].
4. Heat-Shock Protein (HSP) Chaperone System. Misfolded proteins can refold with the assistance of HSP chaperones [8].
5. Integrated Stress Response (ISR). Four serine/threonine protein kinases: (i) GCN2 (nutrient deprivation), (ii) PKR (viral infection), (iii) HRI (heme deprivation), and (iv) PERK (ER stress) are activated by cellular stress. The kinases converge on the phosphorylation of eIF2α [10][11].
6. Unfolded Protein Response (UPR). Misfolded proteins are retained in the ER and delivered for proteasome degradation after retro-translocation into the cytosol [12].
The accumulation of misfolded proteins causes ER stress and activates the UPR via three transducers: (i) activating transcription factor 6 (ATF6), (ii) inositol-requiring enzyme 1 (IRE1), and (iii) protein kinase RNA-like ER kinase (PERK) [12].
3. Translation of drugs targeting proteotoxic stress in cancer – haematologic vs solid cancers
3.1. Exploiting Proteotoxic Stress in Haematologic Malignancies: Multiple Myeloma (MM)
1. Proteasome Inhibitors (PIs). PIs are the mainstay of MM therapy. Proteasome inhibition exploits the unique biology of MM as myeloma cells have large amounts of misfolded proteins, and hence, high levels of proteotoxic stress due to extensive immunoglobulin synthesis [13][14]. There are 3 classes of PIs: (i) boronates, (ii) epoxyketones, and (iii) γ-lactam-β-lactones (salinosporamide).
2. Autophagy Inhibitors. Dual inhibition of autophagy and UPS, with hydroxychloroquine (HCQ) and bortezomib, has recently emerged as a way to overcome therapeutic resistance to proteasome inhibition [15].
3. Aggresomal Pathway Inhibitors. Panobinostat is a broad-spectrum histone deacetylase (HDAC) inhibitors that has been approved for use in refractory/relapsed MM in combination with bortezomib and dexamethasone [16].
4. Heat Shock Protein 90 (HSP90) Inhibitors. HSP90 inhibitors in preclinical development include NVP-HSP990 and TAS-116 [17][18]. Phase I/II trial of tanespimycin (HSP90 inhibitor) in combination with bortezomib has shown a 27% objective response rate (ORR) [19]. However, further development of tanespimycin was halted due to patent expiry [20].
5. Unfolded Protein Response (UPR) Modulators. UPR is activated by drugs (ER stressors) such as tunicamycin, thapsigargin, and brefeldin A. However, these drugs have limited clinical translation due to their anticipated toxicities based on in-vivo studies [21][22][23]. It has been demonstrated that Nelfinavir, a HIV protease inhibitor, potentiates the anti-MM activity of bortezomib via the activation of the UPR and CHOP-dependent apoptosis [24].
3.2. Exploiting Proteotoxic Stress in Solid Tumors: Triple Negative Breast Cancer (TNBC)
Proteasome Inhibition in TNBC. The effect of proteasome inhibitors against solid tumours like triple negative breast cancer (TNBC) has been quite discouraging. A recent study has demonstrated that inhibition of both the β5 and β2 sites of the proteasome sensitised TNBCcell lines to bortezomib [25]. Paclitaxel, a chemotherapeutic drug, has been shown to regulate the genes involved in the ubiquitin proteasome system in breast cancer. Paclitaxel treatment resulted in an upregulation of proteasome genes and lead to an accumulation of proteasome subunits alpha 4 and beta 1, 26S ATPases 2 and 6 and 26S non-ATPase 14 [26].
References
- Richard I. Morimoto; Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & Development 2008, 22, 1427-1438, 10.1101/gad.1657108.
- Ulrich Schubert; Luis C. Antón; James Gibbs; Christopher C. Norbury; Jonathan W. Yewdell; Jack R. Bennink; Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. news@nature 2000, 404, 770-774, 10.1038/35008096.
- Cenci, S.; Sitia, R.; Managing and exploiting stress in the antibody factory. FEBS Lett 2007, 581, 3652–3657, 10.1016/j.febslet.2007.04.031. .
- Davide Ruggero; Translational Control in Cancer Etiology. Cold Spring Harbor Perspectives in Biology 2012, 4, a015891-a015891, 10.1101/cshperspect.a015891.
- Protein quality control and cancerogenesis.. Klinicka Onkologie 2012, 25, 2S38-44, .
- Maisha Chowdhury; Cordula Enenkel; Intracellular Dynamics of the Ubiquitin-Proteasome-System. F1000Research 2015, 4, 367, 10.12688/f1000research.6835.2.
- The Ubiquitin Proteasome Pathway (UPP) . Boston Biochem. Retrieved 2019-2-6
- Tara Lin; Alan K. Ikeda; Tiffany Simms-Waldrip; Cecilia Fu; Kathleen M. Sakamoto; Agustin Rodriguez-Gonzalez; Role of the Aggresome Pathway in Cancer: Targeting Histone Deacetylase 6–Dependent Protein Degradation. Cancer Research 2008, 68, 2557-2560, 10.1158/0008-5472.can-07-5989.
- Daniel J. Metcalf; Moisés García-Arencibia; Warren E. Hochfeld; David C. Rubinsztein; Autophagy and misfolded proteins in neurodegeneration. Experimental Neurology 2012, 238, 22-28, 10.1016/j.expneurol.2010.11.003.
- David J. McConkey; The integrated stress response and proteotoxicity in cancer therapy. Biochemical and Biophysical Research Communications 2017, 482, 450-453, 10.1016/j.bbrc.2016.11.047.
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M; The integrated stress response. EMBO Rep 2017, 482, 450-453, 10.15252/embr.201642195.
- Peter Walter; David Ron; The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science 2011, 334, 1081-1086, 10.1126/science.1209038.
- Raymond J Deshaies; Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biology 2014, 12, 425, 10.1186/s12915-014-0094-0.
- Giada Bianchi; Laura Oliva; Paolo Cascio; Niccolò Pengo; Francesca Fontana; Fulvia Cerruti; Andrea Orsi; Elena Pasqualetto; Alexandre Mezghrani; Valeria Calbi; et al. The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood 2009, 113, 3040-3049, 10.1182/blood-2008-08-172734.
- Dan T Vogl; Edward A Stadtmauer; Kay-See Tan; Daniel F Heitjan; Lisa E Davis; Laura Pontiggia; Reshma Rangwala; Shengfu Piao; Yunyoung C Chang; Emma C Scott; et al. Combined autophagy and proteasome inhibition. Autophagy 2014, 10, 1380-1390, 10.4161/auto.29264.
- Laubach, J.P.; Moreau, P.; San-Miguel, J.F.; Richardson, P.G; Panobinostat for the treatment of multiple myeloma. Clin. Cancer Res 2015, 21, 4767–4773, 10.1158/1078-0432.CCR-15-0530.
- T. Khong; A. Spencer; Targeting HSP 90 Induces Apoptosis and Inhibits Critical Survival and Proliferation Pathways in Multiple Myeloma. Molecular Cancer Therapeutics 2011, 10, 1909-1917, 10.1158/1535-7163.mct-11-0174.
- Suzuki, R.; Hideshima, T.; Mimura, N.; Minami, J.; Ohguchi, H.; Kikuchi, S.; Yoshida, Y.; Gorgun, G.; Cirstea, D.; Cottini, F.; et al. Anti-tumor activities of selective HSP90alpha/beta inhibitor, TAS-116, in combination with bortezomib in multiple myeloma. Leukemia 2015, 29, 510-514, 10.1038/leu.2014.300.
- Paul G. Richardson; Asher A. Chanan-Khan; Sagar Lonial; Amrita Y. Krishnan; Michael P. Carroll; Melissa Alsina; Maher Albitar; David Berman; Marianne Messina; Kenneth C. Anderson; et al. Tanespimycin and bortezomib combination treatment in patients with relapsed or relapsed and refractory multiple myeloma: results of a phase 1/2 study. British Journal of Haematology 2011, 153, 729-740, 10.1111/j.1365-2141.2011.08664.x.
- Bristol-Myers Squibb Halts Development of Tanespimycin . Staff, T.M.B. Retrieved 2019-2-6
- H.-R. Park; A. Tomida; S. Sato; Y. Tsukumo; J. Yun; T. Yamori; Y. Hayakawa; T. Tsuruo; K. Shin-Ya; Effect on Tumor Cells of Blocking Survival Response to Glucose Deprivation. JNCI Journal of the National Cancer Institute 2004, 96, 1300-1310, 10.1093/jnci/djh243.
- Q. Wang; H. Mora-Jensen; M. A. Weniger; P. Perez-Galan; C. Wolford; T. Hai; D. Ron; W. Chen; W. Trenkle; A. Wiestner; et al. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proceedings of the National Academy of Sciences 2009, 106, 2200-2205, 10.1073/pnas.0807611106.
- Danyelle M. Townsend; Yefim Manevich; Lin He; Ying Xiong; Robert R. Bowers; Steven Hutchens; Kenneth D. Tew; Nitrosative Stress-Induced S-Glutathionylation of Protein Disulfide Isomerase Leads to Activation of the Unfolded Protein Response. Cancer Research 2009, 69, 7626-7634, 10.1158/0008-5472.CAN-09-0493.
- M Krauß; J Bader; H Overkleeft; C Driessen; Nelfinavir augments proteasome inhibition by bortezomib in myeloma cells and overcomes bortezomib and carfilzomib resistance. Blood Cancer Journal 2013, 3, e103, 10.1038/bcj.2013.2.
- Emily S. Weyburne; Owen M. Wilkins; Zhe Sha; David A. Williams; Alexandre A. Pletnev; Gerjan De Bruin; Hermann S. Overkleeft; Alfred L. Goldberg; Michael D. Cole; Alexei F. Kisselev; et al. Inhibition of the Proteasome β2 Site Sensitizes Triple-Negative Breast Cancer Cells to β5 Inhibitors and Suppresses Nrf1 Activation. Cell Chemical Biology 2017, 24, 218-230, 10.1016/j.chembiol.2016.12.016.
- Carolina Sánchez-Estévez; Mercedes Julián-Tendero; Héctor Hernández-Vargas; Cayetano Von Kobbe; José Palacios; Gema Moreno-Bueno; Inhibition of Paclitaxel-Induced Proteasome Activation Influences Paclitaxel Cytotoxicity in Breast Cancer Cells in a Sequence-Dependent Manner. Cell Cycle 2007, 6, 2662-2668, 10.4161/cc.6.21.4821.
