Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The mechanism of RNA interference (RNAi) could represent a breakthrough in the therapy of all diseases that arise from a gene defect or require the inhibition of a specific gene expression. In particular, small interfering RNA (siRNA) offers an attractive opportunity to achieve a new milestone in the therapy of human diseases.
- siRNA
- gene delivery
- surface-functionalized nanoparticles
1. Introduction
After the discovery of RNA interference (RNAi) in 1998 [1], two decades had to pass before the full potential of siRNA therapeutics was recognized and the first drug based on this technology, called Onpattro (patisiran, ALNTTR02), found therapeutic application in the clinic in 2018 [2].
The history of the discovery of RNAi already started in 1990, when Napoli et al. tried to increase the expression of the CHS gene in petunias to achieve a stronger flower pigmentation by adding additional copies of the CHS gene into the plants. Contrary to their predictions, the colour faded due to an alleviated pigment formation in the genetically modified plants. The phenomenon was named “co-suppression” because the additional genes also reduced the expression of the naturally occurring CHS gene [3]. Further research showed that the inhibition of the genes does not take place at transcription level and that increased formed mRNA degrades rapidly. This effect was called “Post-Transcriptional Gene Silencing” (PTGS) [4]. Finally, in 1998, it was demonstrated by Fire et al. in the nematode “Caenorhabditis elegans” that the produced mRNA itself is involved in PTGS and that efficient and specific gene knockdown is possibly caused by the delivery of double-stranded RNA [1]. Based on their pioneering findings, Andrew Fire and Craig Mello were awarded with the Nobel Prize in Physiology or Medicine in 2006 for the discovery of RNA interference [5]. In summary, starting with the silencing of genes in plants in 1990 [3] and in nematodes in 1998 [1], the technique evolved further, leading to the silencing of genes in different mammalian cell lines in 2001 [6]. Currently it is possible to block almost any gene of interest in humans by double-stranded RNA sequences or the significant smaller small-interfering RNA (siRNA), resulting in tremendous potential for RNAi usage [7][8][9].
Explained briefly, siRNA is able to abrogate the expression of selected genes through the mechanism of RNAi [10]. The potential in RNAi is impressive, since it is a highly guarded and conserved mechanism that can be found in eukaryotic cells, where it serves as a natural defender against foreign and potential dangerous nucleic acids entering the cell [7][11]. Furthermore, the process is so efficient that about 2000 siRNA molecules per cell are sufficient for an efficient response. Using nucleic acids for “silencing” genes comes with benefits. It is convenient for the development of new drugs, as it enables a re-targeting without big alterations of the drug formulation. Theoretically, the site of action and the pharmacological effect of the drug might be “customized”, switching off the gene of interest by replacing the siRNA sequence [11]. Furthermore, siRNA can easily and reproducibly be synthesized at large scale.
Multiple disadvantages and barriers in the handling and application of siRNA such as poor stability, rapid degradation by enzymes, low uptake into the desired target tissue, and insufficient encapsulation efficacy as well as effectiveness in transfection were overcome by new findings and advances in the application of siRNA through the use of appropriate surface functionalized drug delivery systems [9].
In order to precise the application of nanoparticles at the desired tissue, it makes sense to target molecular structures through the surface functionalization of nanoparticulate systems [12]. With further attention to development and research, the probability that the product will reach the daily clinical practice will strongly increase. In addition, the efficacy of nanocarrier-based drug delivery systems mainly depends on their controlled interactions with biomolecules. To enhance the efficacy of NPs and increase the delivery rate to targeted tissues, surface modification of particles is a promising option [8][12].
2. Silencing of Genes: Fate of the siRNA
Gene therapy enables a versatile way for patients with life threatening diseases to get a treatment that better suits the individual and works in a more specific fashion. With the delivery of nucleic acids, the effects of genes may not only be turned off but can also be augmented or changed in the desired tissues. On the one hand, genes can be inhibited using small interfering RNA (siRNA), microRNA (miRNA) and inhibitory antisense oligonucleotides (ASOs) [13], which can silence the translation of the protein. On the other hand, DNA plasmids or messenger RNA (mRNA) can be introduced into the cell to promote or alter the expression of genes resulting in an increased synthesis of the targeted protein [14]. Many of the above-mentioned methods are applicable therapeutically, but researchers would like to take a deeper look at the system of RNA interference.
2.1. Challenges and Barriers for siRNA
Following intravenous application, siRNA is circulating in the bloodstream. In order to be effective, siRNA must leave the bloodstream and cross the cell membrane to enter the cytosol of the cell in an intact manner by overcoming the endosomal/lysosomal compartment [15]. Notably, this turns out to be difficult for the circulating siRNA due to its physical and chemical properties, since both the siRNA, as well as the cell membrane, are negatively charged, leading to a repulsion between the RNA and the membrane [7]. In addition, nonspecific bindings by plasma proteins further complicate the distribution of siRNA to its target site. Furthermore, the size of siRNA with a length less than 8 nm and a diameter smaller than 3 nm is still too large to penetrate the cell membrane directly [16]. However, the size is small enough to be excreted within a few minutes glomerularly by the kidney and to accumulate in the urine, which is another disadvantageous factor with regard to effectiveness [17][18]. In addition to the rapid clearance in the bloodstream, siRNA is exposed to RNAses in the plasma and tissue. Nucleases can cut the siRNA rapidly into inactive fragments with a higher efficiency than for DNA due to the 2′-hydroxy group of the RNA. siRNA is a target for defence mechanisms of the host, not only extracellularly but also intracellularly. Thus, siRNA taken up by endocytosis must first escape out of the endosomal and lysosomal vesicles to act in the cytosol [19]. In this context, siRNA packed into nanoparticles might be beneficial, enabling endosomal release via the proton sponge effect or preventing endosomal uptake of the siRNA in general via fusion or membrane penetration directly [19][20].
2.2. Mechanism of Action within the Cell
After the siRNA successfully managed the transport into the cell, the mechanism of RNAi as a natural defence mechanism against potential dangerous nucleic acids is activated, as shown in Figure 1 [21]. There is no difference whether a short sequence of siRNA or a longer double stranded RNA (dsRNA) is detected in the cytosol of the cell, as longer dsRNA is cut by the endoribonuclease dicer into small 20–25 base pair siRNA sequences [22][23].
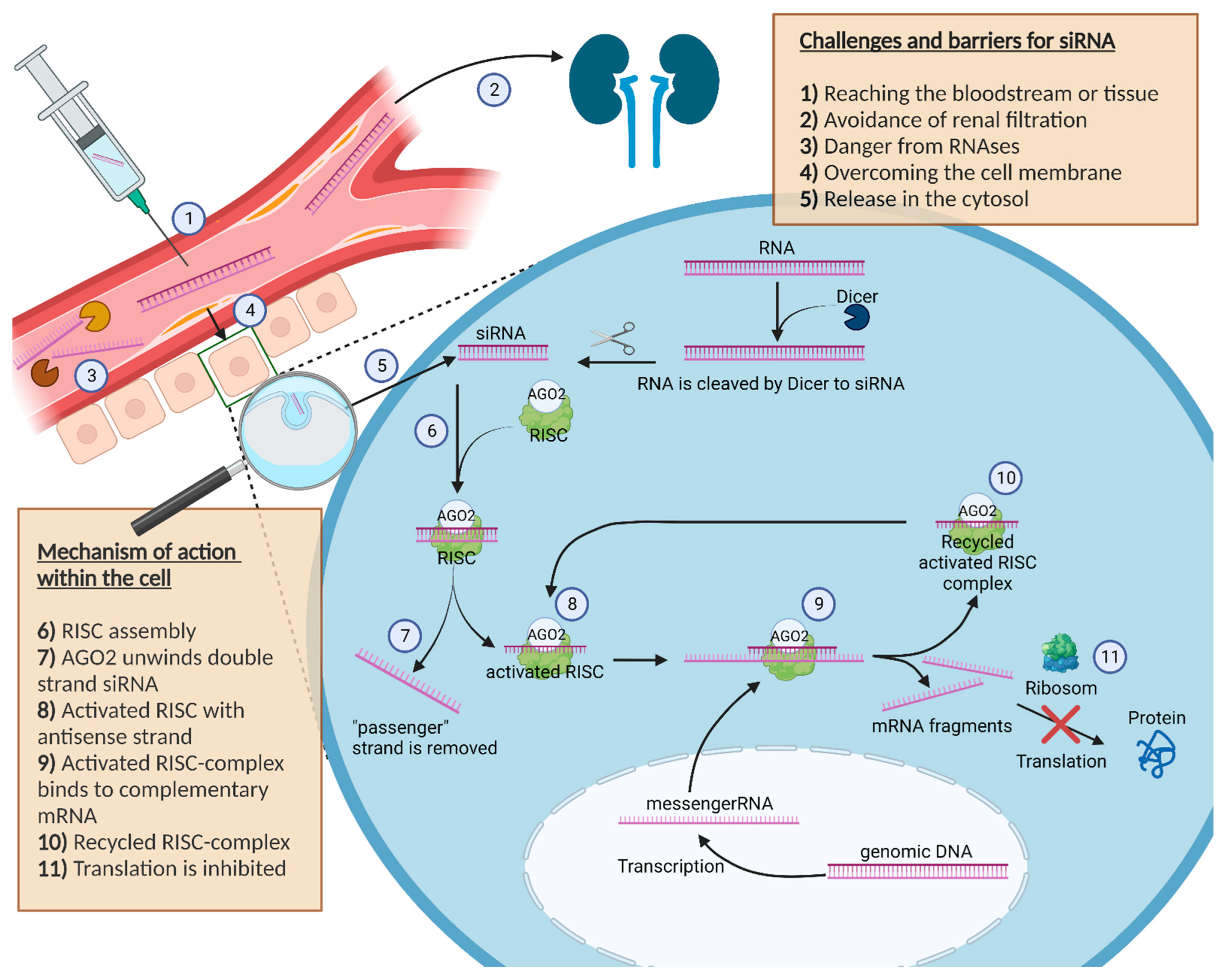
Figure 1. Schematic illustration about the challenges, barriers and the mechanism of action of siRNA in the cell. Furthermore, the figure shows the journey of siRNA starting at the injection site to the cell cytosol, where siRNA can exert its pharmacological effect. The description is in the box inside the picture.
Unlike a longer piece of dsRNA, the direct introduction of siRNA avoids a possible innate immune reaction with interferons caused by the interaction with intracellular receptors or by the activity of the enzyme dicer itself [24]. In the cytoplasm, double stranded siRNA binds to the RNA-induced silencing complex (RISC). A further component of the RISC complex called Argonaut 2 (AGO2) unwraps the double stranded siRNA in a single siRNA strand and removes the unnecessary “passenger” strand [25]. The activated RISC complex contains the antisense strand of the siRNA and guides the single-stranded antisense RNA inside the complex to the matching complementary mRNAs of the cell [26].
If base pairing between the antisense strand inside the activated RISC complex and the complementary mRNA strand matches, the RISC complex cleaves the mRNA and the fragments get degraded. As a result, the initial protein can no longer be translated [22][23][26].
After cleaving the target mRNA, the RISC complex with the included single-stranded siRNA can slice additional molecules of the target mRNA [27].
Thus, with only a few siRNA molecules in the cell cytosol, it is possible to slice an enlarged number of mRNA molecules within the cell, which represents the amplifying effect of the RISC complex [15], highlighting why the used siRNA has no stochiometric ratio to the degraded mRNA in the cell. The RISC mechanism further explains why it is possible to reliably silence genes by siRNA.
Bartlett et al. outlined that the silencing effect caused by siRNA lasts in rapidly dividing cells for several days and in slow dividing cells for up to several weeks. The activated RISC complex is stable for weeks; however, its concentration decreases with each cell division [28].
Showing that the amount of administered siRNA exerts a significant effect on the magnitude of gene silencing, but only a small effect on the duration of it, Bartlett et al. transfected non-dividing cells with four different siRNA concentrations (10 nM, 25 nM, 50 nM, 100 nM). They noticed significant differences in the magnitude of gene silencing, but only small alterations in the gene silencing duration. In addition, to show that the division rate of the cells has a significant impact on the gene knockdown duration, they used 4 different cell lines with different doubling times: Neuro2A-Luc (0.8 days), LNCaP-Luc (1.4 days), HeLa-Luc (1.6 days) and CCD-1074Sk-Luc (non-dividing). They demonstrated that cells with a high doubling rate had a shorter duration of gene silencing due to a dilution effect resulting from cell division [28].
2.3. siRNA in Function of a Drug: Effects and Side Effects
Using siRNA in a function like a drug has further advantages. Contrary to small molecular drugs or monoclonal antibodies, siRNA needs a full complementary Watson-crick base pairing, in other words a perfect nucleotide base pairing, to their matching opposite mRNA to exert the pharmacological effect. This makes the mechanism of siRNA very specific. In contrast, small molecular drugs must be able to recognize the complex spatial arrangement of their target, and moreover antibodies must be able to recognize the epitope of the active target structure in order to be effective. Unfortunately, not every disorder can be treated with small molecular drugs or monoclonal antibodies to date considering the lack of molecular targets [7].
At the same time, it has to be mentioned that with a few nucleotide mismatches at the complementary base pairing, the risk of off-target effects of siRNA increases, although single mismatched base pairings are expected to be tolerated by the RISC complex. In the case of larger contiguous mismatched base pairs, however, unspecific off-target effects can occur such as interferon immune responses by activation of Toll-like receptor 3 (TLR3) or TLR7 [29]. TLR3 activation can cause cell apoptosis, as Cho et al. showed in a neovascularization mouse model, in which non-targeted siRNA suppresses hemangiogenesis and lymphangiogenesis [30]. Furthermore, there may be a change in the expression of other undesired genes leading to non-specific effects [31]. Additionally, an unintentional knockdown of other genes can occur if the base pairing coincidentally matches the sequence of the siRNA incorporated in the RISC complex, resulting in degradation of the matched mRNA [32]. It seems that anywhere from 7 to 11 mismatched contiguous base pairings in siRNA can induce the translation repression of mRNA [32][33]. Nevertheless, siRNA is highly specific for effective gene silencing and has predominantly only one mRNA target, therefore, it is suitable for an effective suppression of mRNA translation.
Significant progress was made in the prevention from off-target effects and optimizing the safety of siRNA utilization. This includes the selection of sequences having only marginal overlap in the nucleotide matching with other mRNA sequences, or the siRNA structure can be modified chemically [34][35]. One example for an effective way to avoid off-target effects can be achieved by using siRNA bundles. SiRNA bundles are a whole series of different enzymatically generated siRNA sequences with the same target gene. The use of siRNA bundles shows a very potent knockdown without showing detectable off-target effects [36].
Despite the enormous therapeutic potential of siRNA and the growing interest in this technology, there are only a few siRNA-based drugs that are approved for clinical application [37][38][39] or under clinical investigation [40][41][42]. In conclusion, with all the difficult drug properties of siRNA, the administration of blank siRNA is insufficient. To improve the pharmacological potential of siRNA, several nanoparticulate systems have been evolved which will be discussed further in the following section.
3. Types of Nanoparticulate Drug Delivery Systems
Nanotechnologies and their possible application in medical fields such as therapy and diagnostics but also in electronics, material sciences and chemistry have recently attracted more and more interest from both scientists and non-scientists. For the pharmaceutical development of new therapies, nanoparticles with a size ≤100 nm are required because they offer different and diverse aspects in their structure and characteristics due to their large surface area [43][44][45]. As mentioned previously, siRNA has to overcome a lot of barriers and hazards in order to fulfil the pharmacological effect in the cell. Therefore, researchers developed various techniques and formulations to encapsulate the siRNA to counteract barriers and hazards. Kim et al. highlighted the current trends for siRNA drug delivery platforms [46]. They outlined that lipid-based nanoparticles followed by polymeric-based nanoparticles are superior to other drug delivery systems, such as metal- (gold and iron oxide) or silicon-based systems. Consequently, this research will focus on the possible surface design options in lipid- and polymer-based nanoparticles, as they are the leading drug delivery systems for siRNA.
The formulation and application strategies of many nanoscaled drug delivery systems were adapted from DNA to RNA. Mechanistic analogies can be found between the two nucleic acids in the process of complexation due to the negative charge, protection from DNAses or RNAses, as well as the release of the nucleic acids into the cell, since in both cases DNA and RNA are double-stranded nucleic acids with an anionic phosphodiester backbone [47]. However, differences already become obvious when looking at the size, structure and chemistry of DNA and siRNA molecules. The weight of plasmid DNA with up to several thousand base pairs is several times larger than the weight of siRNA with about 20–25 base pairs, and therefore shows significantly more electrostatic interactions with polycations, which is an advantage for the nanoparticle formation of DNA. Likewise, the DNA backbone shows a significant improvement in stability due to the stable deoxyribose component of DNA, in contrast to the sensitive 2′-hydroxy group of ribose on the RNA backbone [7].
Nevertheless, there are already quite a few ways to stabilize siRNA by chemical modifications, such as the introduction of 2′-fluorine or 2′-methoxy groups, thus making the application of siRNA more feasible [48]. Considering the site of action of the two nucleic acids, DNA must be transported into the nucleus, whereas siRNA already reached its target in the cytosol. Therefore, a suitable transport vehicle for siRNA has the focus on a reliable release from the endosome into the cytosol. Additional properties, such as buffer capacities at endosomal pH values or possible lytic properties that allow an enhanced nucleic acid release from the endosome, offer advantages [49].
However, due to the higher instability of RNA resulting from the 2′-hydroxy group, the lower chain length and the number of anionic charges, a less efficient electrostatic interaction as well as nanoparticle formation occurs. Therefore, a direct transfer from DNA to RNA is not possible, and an adaption of the formulation processes is necessary.
3.1. Lipid-Based Nanoparticles
Lipid-based drug delivery systems are the most widely used and marketed systems that include a whole range of carrier types. The most advanced liposomes are large amphiphilic vesicles with a lipid bilayer having both a polar aqueous core and an unpolar surrounding lipid bilayer. More complex lipid nanoparticles such as solid lipid nanoparticles, nanostructured lipid carriers and electrostatic cationic lipid- nucleic acid lipoplexes have a more complex structure and enhanced properties in stability compared to liposomes [50][51][52][53]. Originally, LNPs were developed as vehicles for DNA-based drugs. Over time, they have proven to be reliable delivery systems for siRNA, as they are also able to entrap siRNA and thus protect the siRNA from degradation. The lipid matrix of the majority of siRNA LNPs is composed of cationic or ionizable lipids, neutral helper lipids, cholesterol and shielding lipids [54].
Cationic lipids such as DOTAP [55], DOTMA [56] or DMAPAP [57] can electrostatically form stable complexes with the negative siRNA due to electrostatic interactions, thereby enabling the capturing of siRNA in the LNP. However, the net cationic lipid systems suffer from the so called “polycationic dilemma”, as a high number of cationic charges favour the formation of complexes, but vice versa suffer from cytotoxic effects due to unspecific interactions with negatively charged cell membranes and blood components [58]. In that regard, research about pH-sensitive ionizable lipids was intensified [59]. Ionizable lipids such as DLin-DMA, DODAP or DLin-MC3-DMA offer a key advantage during the production of LNPs, because their charge can be varied [55]. They can be positively or neutrally charged depending on the pH value of the medium and allow reliable electrostatic complexation with siRNA, resulting in efficient encapsulation during manufacturing. This ability to change the charge at different pH values reduces the toxicity of ionizable lipids [60]. On the other hand, at acidic conditions in the endosomal/lysosomal compartments, ionizable lipids can use the ability to alter their charge and interact with negatively charged cell structures to destabilize the endosomal membrane and thereby promote the release of siRNA into the cell cytosol [61].
The contribution of helper lipids such as DSPC and cholesterol seems to be crucial for the stability of LNPs. A typical helper lipid that is widely used in siRNA LNPs is DSPC [62]; however, the mechanism of action has yet to be fully understood. The helper lipid is normally used in low molar concentrations in the LNP formulation and is presumed to interact with the lipid layer, thus stabilizing the LNP [63]. Another component that has a significant effect on lipid packing, membrane fluidity and permeability of the lipid bilayer is cholesterol [64]. By inserting cholesterol in the lipid formulation, it is possible to reduce the distance between the individual lipids in the lipid layer and thereby prevent the premature release of the active ingredient [65].
In addition, the characteristics of shielding lipids are interesting as well. Shielding lipids are often associated with the biocompatible hydrophilic macromolecular polymer poly (ethylene glycol) (PEG), which was initially invented to hide and protect biological substances due to the fact that they can be removed rapidly from the bloodstream by detection from macrophages [66]. PEG alternatives such as synthetic polymers (e.g., poly (glycerine), poly (acrylamide)), natural polymers (e.g., polyamino acids, glycosaminoglycans) or zwitterionic polymers (e.g., poly (carboxybetaine), poly (sulfobetaine)) are currently under intensive investigation due to PEG side effects such as the ABC phenomenon [67], the formation of PEG antibodies, as well as a low drug efficacy as a consequence of anti-PEG antibodies [68][69]. Further shielding of lipids increases the circulation half-life and stability and prevents the aggregation of particles during storage [70]. However, when using PEG, the so-called “PEG dilemma” is also encountered, whereby the efficient transfection of cells may be compromised because PEG shielding may not only protect the LNPs from the recognition by the immune system but also from the interaction with target cells [71]. As a consequence, the reduced transfection efficiency by PEGylation can be improved using special PEG lipids, where the linkage between PEG and lipid opens after administration time-dependent or in a stimuli-responsive manner and exposes the LNP surface, making them available for cell transfection once again [72]. Nevertheless, since the most common administration of LNPs is an intravenous injection into the body, resulting in a rapid systemic distribution, LNPs suffer from a short half-life when administered parenterally. To ensure a controlled or long half-life, the assistance of further delivery systems, such as hydrogels, for example, is recommended [73]. This allows the use of additional routes of administration to protect the LNPs from rapid clearance and to release it in a controlled manner from a drug depot at the target tissue [74]. Concluding LNPs achieve a high loading capacity for siRNA as well as an efficient transfection of cells; however, in the case of prolonged or delayed release, they are limited.
3.2. Polymer-Based Nanoparticles
Polymer NPs are the other main class for siRNA delivery. They represent a wide and diverse fraction of matrices with variations of each formulation regarding the type of encapsulation (nanospheres and nanocapsules). Polymers offer a reliable and proven way to control the release of drugs through their method of application (oral, parenteral, subcutaneous, local or systemic) and can be modified for sustained and controlled release [75][76]. Through the selection of various polymers in the carrier matrix, several different release mechanisms can be applied (diffusion- and solvent -controlled release, polymer-degraded or pH-sensitive release) [77]. This allows to influence the drug release and to adjust a therapeutic range and can prevent multiple drug administrations to the patient. At the same time, polymer NP offers high physicochemical stability, various targeting moieties through their diverse modification capabilities, as well as good transfection efficiency [78]. By using polymers, many formulations can be established, such as poly (lactid-co-glycolid) (PLGA) [79], poly (L-lysine) (PLL) [80], poly (L-arginine) (PLA) [81], poly (methacrylamide) [82], chitosan [83], poly (ethylene imine) (PEI) [84], cyclodextrin [85], hyaluronic acid [86], gelatine, [87] and alginate [88], which represent a selection of different polymers used in drug delivery systems.
These polymers can be divided by natural and synthetic origin. Thus, in the group of natural polymers, candidates can be found, such as polysaccharides [89], peptides [90], or bacterial nanocellulose [91]. The group of synthetic polymers includes representatives such as PLGA or poly (lactic acid) (PLA), amongst others [92]. Both types have their pros and cons. While natural polymers stand out due to their good biocompatibility and material availability, synthetic polymers often impress with their enormous variations and the possibility to produce them precisely and in a purified manner. The high purity of natural polymers is more challenging to achieve, as natural by-products may be present. The major disadvantage of synthetic polymers is their often problematic biocompatibility and biodegradability [93].
PLGA, for example, as a widely used polymer in drug delivery is already marketed in several applications, and is often adapted as a particle matrix. Due to its good biocompatibility and biodegradability, and potential to control drug release, it is applied in formulations approved by regulatory authorities [94][95]. However, the encapsulation of the water-soluble siRNA in the lipophilic PLGA is challenging. During production, siRNA can escape due to its hydrophilic properties and electrostatic repulsions, which are caused by the phosphate groups in the siRNA and the carboxylic residues of the PLGA polymer. As a result, only a small amount of siRNA can be entrapped inside the PLGA particles [95]. The use of cationic polymers such as PEI or oligopeptides (such as poly (beta-amino ester) to complex the nucleic acids into the polymer matrix can increase the entrapment efficiency [96][97].
Another innovative approach is to combine the advantages of both polymers and lipids, as shown by Ewe et al. [98]. By combining PEI/siRNA polyplexes with different phospholipid liposomes (consisting of the lipids DPPC, DPPC/DPPE or DPPC/DPPG), so called “lipopolyplexes” can be generated. These enable PEI to efficiently condense nucleic acids and at the same time to mask the cytotoxicity of the PEI polymer through good biocompatible properties of the lipids. In an ex vivo PC-3 prostate carcinoma xenograft model, both PEI/siRNA polyplexes and PEI/siRNA/DPPC lipopolyplexes were able to silence 50% of the survivin gene [98]. To summarize, in this study the benefit of polymers and lipids in combination provided a high knockdown efficiency with good biocompatibility.
This entry is adapted from the peer-reviewed paper 10.3390/ijms232213929
References
- Fire, A.; Xu, S.Q.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811.
- Weng, Y.H.; Xiao, H.H.; Zhang, J.C.; Liang, X.J.; Huang, Y.Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019, 37, 801–825.
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in Trans. Plant Cell 1990, 2, 279–289.
- Vanblokland, R.; Vandergeest, N.; Mol, J.N.M.; Kooter, J.M. Transgene-Mediated Suppression of Chalcone Synthase Expression in Petunia-Hybrida Results from an Increase in Rna Turnover. Plant J. 1994, 6, 861–877.
- Bernards, R. The Nobel Prize in Physiology or Medicine for 2006 for the discovery of RNA interference. Ned. Tijdschr. Geneeskd. 2006, 150, 2849–2853.
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498.
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target Ther. 2020, 5, 101.
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147.
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444.
- Geley, S.; Muller, C. RNAi: Ancient mechanism with a promising future. Exp. Gerontol. 2004, 39, 985–998.
- Setten, R.L.; Rossi, J.J.; Han, S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446.
- Sanita, G.; Carrese, B.; Lamberti, A. Nanoparticle Surface Functionalization: How to Improve Biocompatibility and Cellular Internalization. Front. Mol. Biosci. 2020, 7, 587012.
- Schoch, K.M.; Miller, T.M. Antisense Oligonucleotides: Translation from Mouse Models to Human Neurodegenerative Diseases. Neuron 2017, 94, 1056–1070.
- Weng, Y.H.; Li, C.H.; Yang, T.R.; Hu, B.; Zhang, M.J.; Guo, S.; Xiao, H.H.; Liang, X.J.; Huang, Y.Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020, 40, 107534.
- Wittrup, A.; Ai, A.; Liu, X.; Hamar, P.; Trifonova, R.; Charisse, K.; Manoharan, M.; Kirchhausen, T.; Lieberman, J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015, 33, 870–876.
- Wu, L.; Zhou, W.; Lin, L.; Chen, A.; Feng, J.; Qu, X.; Zhang, H.; Yue, J. Delivery of therapeutic oligonucleotides in nanoscale. Bioact. Mater. 2022, 7, 292–323.
- Park, J.; Park, J.; Pei, Y.; Xu, J.; Yeo, Y. Pharmacokinetics and biodistribution of recently-developed siRNA nanomedicines. Adv. Drug Deliv. Rev. 2016, 104, 93–109.
- Longmire, M.; Choyke, P.L.; Kobayashi, H. Clearance properties of nano-sized particles and molecules as imaging agents: Considerations and caveats. Nanomedicine 2008, 3, 703–717.
- Wittrup, A.; Lieberman, J. Knocking down disease: A progress report on siRNA therapeutics. Nat. Rev. Genet. 2015, 16, 543–552.
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol.. 2015, 33, 941–951.
- Gaynor, J.W.; Campbell, B.J.; Cosstick, R. RNA interference: A chemist’s perspective. Chem. Soc. Rev. 2010, 39, 4169–4184.
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366.
- Nakanishi, K. Anatomy of RISC: How do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip. Rev. RNA 2016, 7, 637–660.
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006, 441, 537–541.
- Rand, T.A.; Ginalski, K.; Grishin, N.V.; Wang, X.D. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. USA 2004, 101, 14385–14389.
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther.-Nucl. Acids 2015, 4, e252.
- Hutvagner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060.
- Bartlett, D.W.; Davis, M.E. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006, 34, 322–333.
- Hornung, V.; Guenthner-Biller, M.; Bourquin, C.; Ablasser, A.; Schlee, M.; Uematsu, S.; Noronha, A.; Manoharan, M.; Akira, S.; de Fougerolles, A.; et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005, 11, 263–270.
- Cho, W.G.; Albuquerque, R.J.C.; Kleinman, M.E.; Tarallo, V.; Greco, A.; Nozaki, M.; Green, M.G.; Baffi, J.Z.; Ambati, B.K.; De Falco, M.; et al. Small interfering RNA-induced TLR3 activation inhibits blood and lymphatic vessel growth. Proc. Natl. Acad. Sci. USA 2009, 106, 7137–7142.
- Scacheri, P.C.; Rozenblatt-Rosen, O.; Caplen, N.J.; Wolfsberg, T.G.; Umayam, L.; Lee, J.C.; Hughes, C.M.; Shanmugam, K.S.; Bhattacharjee, A.; Meyerson, M.; et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1892–1897.
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637.
- Chen, J.S.; Peng, Y.C.; Zhang, H.N.; Wang, K.X.; Zhao, C.Q.; Zhu, G.H.; Palli, S.R.; Han, Z.J. Off-target effects of RNAi correlate with the mismatch rate between dsRNA and non-target mRNA. Rna Biol. 2021, 18, 1747–1759.
- Bartoszewski, R.; Sikorski, A.F. Editorial focus: Understanding off-target effects as the key to successful RNAi therapy. Cell Mol. Biol. Lett. 2019, 24, 69.
- Khvorova, A.; Watts, J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017, 35, 238–248.
- Hannus, M.; Beitzinger, M.; Engelmann, J.C.; Weickert, M.T.; Spang, R.; Hannus, S.; Meister, G. siPools: Highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014, 42, 8049–8061.
- Wood, H. FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat. Rev. Neurol. 2018, 14, 570.
- Scott, L.J. Givosiran: First Approval. Drugs 2020, 80, 335–339.
- Scott, L.J.; Keam, S.J. Lumasiran: First Approval. Drugs 2021, 81, 277–282.
- Raal, F.J.; Kallend, D.; Ray, K.K.; Turner, T.; Koenig, W.; Wright, R.S.; Wijngaard, P.L.J.; Curcio, D.; Jaros, M.J.; Leiter, L.A.; et al. Inclisiran for the Treatment of Heterozygous Familial Hypercholesterolemia. N. Engl. J. Med. 2020, 382, 1520–1530.
- Moreno-Montanes, J.; Bleau, A.M.; Jimenez, A.I. Tivanisiran, a novel siRNA for the treatment of dry eye disease. Expert. Opin. Investig. Drugs 2018, 27, 421–426.
- Zhang, M.M.; Bahal, R.; Rasmussen, T.P.; Manautou, J.E.; Zhong, X.B. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol. 2021, 189, 114432.
- Zink, M.; Hotzel, K.; Schubert, U.S.; Heinze, T.; Fischer, D. Amino Acid-Substituted Dextran-Based Non-Viral Vectors for Gene Delivery. Macromol. Biosci. 2019, 19, 1900085.
- Nasr, S.S.; Lee, S.; Thiyagarajan, D.; Boese, A.; Loretz, B.; Lehr, C.M. Co-Delivery of mRNA and pDNA Using Thermally Stabilized Coacervate-Based Core-Shell Nanosystems. Pharmaceutics 2021, 13, 1924.
- Friedrich, B.; Auger, J.P.; Dutz, S.; Cicha, I.; Schreiber, E.; Band, J.; Boccacccini, A.R.; Kronke, G.; Alexiou, C.; Tietze, R. Hydroxyapatite-Coated SPIONs and Their Influence on Cytokine Release. Int. J. Mol. Sci. 2021, 22, 4143.
- Kim, B.; Park, J.H.; Sailor, M.J. Rekindling RNAi Therapy: Materials Design Requirements for In Vivo siRNA Delivery. Adv. Mater. 2019, 31, e1903637.
- Scholz, C.; Wagner, E. Therapeutic plasmid DNA versus siRNA delivery: Common and different tasks for synthetic carriers. J. Control. Release 2012, 161, 554–565.
- Watts, J.K.; Deleavey, G.F.; Damha, M.J. Chemically modified siRNA: Tools and applications. Drug. Discov. Today 2008, 13, 842–855.
- Meyer, M.; Dohmen, C.; Philipp, A.; Kiener, D.; Maiwald, G.; Scheu, C.; Ogris, M.; Wagner, E. Synthesis and Biological Evaluation of a Bioresponsive and Endosomolytic siRNA-Polymer Conjugate. Mol. Pharmaceut. 2009, 6, 752–762.
- Hattori, Y.; Nakamura, M.; Takeuchi, N.; Tamaki, K.; Shimizu, S.; Yoshiike, Y.; Taguchi, M.; Ohno, H.; Ozaki, K.I.; Onishi, H. Effect of cationic lipid in cationic liposomes on siRNA delivery into the lung by intravenous injection of cationic lipoplex. J. Drug Target. 2019, 27, 217–227.
- Viegas, J.S.R.; Praça, F.G.; Caron, A.L.; Suzuki, I.; Silvestrini, A.V.P.; Medina, W.S.G.; Del Ciampo, J.O.; Kravicz, M.; Bentley, M.V.L.B. Nanostructured lipid carrier co-delivering tacrolimus and TNF-α siRNA as an innovate approach to psoriasis. Drug Deliv. Transl Res. 2020, 10, 646–660.
- Francis, J.E.; Skakic, I.; Dekiwadia, C.; Shukla, R.; Taki, A.C.; Walduck, A.; Smooker, P.M. Solid Lipid Nanoparticle Carrier Platform Containing Synthetic TLR4 Agonist Mediates Non-Viral DNA Vaccine Delivery. Vaccines 2020, 8, 551.
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q.Q. Lipid Nanoparticles-From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. Acs Nano. 2021, 15, 16982–17015.
- Kanasty, R.; Dorkin, J.R.; Vegas, A.; Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013, 12, 967–977.
- Yonezawa, S.; Koide, H.; Asai, T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliver. Rev. 2020, 154, 64–78.
- Hou, X.C.; Zaks, T.; Langer, R.; Dong, Y.Z. Lipid nanoparticles for mRNA delivery (Aug, 10./s41578–021–00358–0, 2021). Nat. Rev. Mater. 2022, 7, 65.
- Liu, X.; Bahloul, B.; Lai Kuen, R.; Andrieux, K.; Roques, C.; Scherman, D. Cationic lipid nanoparticle production by microfluidization for siRNA delivery. Int. J. Pharm. 2021, 605, 120772.
- Bernkop-Schnurch, A. Strategies to overcome the polycation dilemma in drug delivery. Adv. Drug Deliv. Rev. 2018, 136, 62–72.
- Semple, S.C.; Klimuk, S.K.; Harasym, T.O.; Dos Santos, N.; Ansell, S.M.; Wong, K.F.; Maurer, N.; Stark, H.; Cullis, P.R.; Hope, M.J.; et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: Formation of novel small multilamellar vesicle structures. Bba-Biomembranes. 2001, 1510, 152–166.
- Belliveau, N.M.; Huft, J.; Lin, P.J.C.; Chen, S.; Leung, A.K.K.; Leaver, T.J.; Wild, A.W.; Lee, J.B.; Taylor, R.J.; Tam, Y.K.; et al. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA. Mol.-Nucl Acids. 2012, 1, e37.
- Hafez, I.M.; Cullis, P.R. Roles of lipid polymorphism in intracellular delivery. Adv. Drug Deliv. Rev. 2001, 47, 139–148.
- Kulkarni, J.A.; Myhre, J.L.; Chen, S.; Tam, Y.Y.C.; Danescu, A.; Richman, J.M.; Cullis, P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomed.-Nanotechnol. 2017, 13, 1377–1387.
- Kulkarni, J.A.; Witzigmann, D.; Leung, J.; Tam, Y.Y.C.; Cullis, P.R. On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale 2019, 11, 21733–21739.
- Briuglia, M.L.; Rotella, C.; McFarlane, A.; Lamprou, D.A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015, 5, 231–242.
- Leeb, F.; Maibaum, L. Spatially Resolving the Condensing Effect of Cholesterol in Lipid Bilayers. Biophys. J. 2018, 115, 2179–2188.
- Savoca, K.V.; Abuchowski, A.; Vanes, T.; Davis, F.F.; Palczuk, N.C. Preparation of a Non-Immunogenic Arginase by the Covalent Attachment of Polyethylene-Glycol. Biochim. Biophys. Acta. 1979, 578, 47–53.
- Thi, T.T.H.; Pilkington, E.H.; Nguyen, D.H.; Lee, J.S.; Park, K.D.; Truong, N.P. The Importance of Poly(ethylene glycol) Alternatives for Overcoming PEG Immunogenicity in Drug Delivery and Bioconjugation. Polymers 2020, 12, 298.
- Abu Lila, A.S.; Ishida, T. Anti-PEG IgM Production via a PEGylated Nanocarrier System for Nucleic Acid Delivery. Methods Mol. Biol. 2019, 1943, 333–346.
- Moreno, A.; Pitoc, G.A.; Ganson, N.J.; Layzer, J.M.; Hershfield, M.S.; Tarantal, A.F.; Sullenger, B.A. Anti-PEG Antibodies Inhibit the Anticoagulant Activity of PEGylated Aptamers. Cell Chem. Biol. 2019, 26, 634.
- Paolino, D.; Accolla, M.L.; Cilurzo, F.; Cristiano, M.C.; Cosco, D.; Castelli, F.; Sarpietro, M.G.; Fresta, M.; Celia, C. Interaction between PEG lipid and DSPE/DSPC phospholipids: An insight of PEGylation degree and kinetics of de-PEGylation. Colloid Surf. B 2017, 155, 266–275.
- Zhao, C.Y.; Deng, H.Z.; Xu, J.; Li, S.Y.; Zhong, L.; Shao, L.H.; Wu, Y.; Liang, X.J. “Sheddable” PEG-lipid to balance the contradiction of PEGylation between long circulation and poor uptake. Nanoscale 2016, 8, 10832–10842.
- Mui, B.L.; Tam, Y.K.; Jayaraman, M.; Ansell, S.M.; Du, X.Y.; Tam, Y.Y.C.; Lin, P.J.C.; Chen, S.; Narayanannair, J.K.; Rajeev, K.G.; et al. Influence of Polyethylene Glycol Lipid Desorption Rates on Pharmacokinetics and Pharmacodynamics of siRNA Lipid Nanoparticles. Mol. Ther.-Nucl. Acids 2013, 2, e139.
- Lyu, D.; Chen, S.; Guo, W. Liposome Crosslinked Polyacrylamide/DNA Hydrogel: A Smart Controlled-Release System for Small Molecular Payloads. Small 2018, 14, e1704039.
- Elkhoury, K.; Kahn, C.; Sanchez-Gonzalez, L.; Arab-Tehrany, E. Liposomes for Biomedical Applications; Soft Matter Series; Azevedo, H.S., Mano, J.F., Borges, J., Eds.; Royal Society of Chemistry: London, UK, 2021; pp. 392–404.
- Shetab Boushehri, M.A.; Dietrich, D.; Lamprecht, A. Nanotechnology as a Platform for the Development of Injectable Parenteral Formulations: A Comprehensive Review of the Know-Hows and State of the Art. Pharmaceutics 2020, 12, 510.
- Ali, H.; Weigmann, B.; Neurath, M.F.; Collnot, E.M.; Windbergs, M.; Lehr, C.M. Budesonide loaded nanoparticles with pH-sensitive coating for improved mucosal targeting in mouse models of inflammatory bowel diseases. J. Control. Release 2014, 183, 167–177.
- Son, G.-H.; Lee, B.-J.; Cho, C.-W. Mechanisms of drug release from advanced drug formulations such as polymeric-based drug-delivery systems and lipid nanoparticles. J. Pharm. Investig. 2017, 47, 287–296.
- Prajapati, S.K.; Jain, A.; Jain, A.; Jain, S. Biodegradable polymers and constructs: A novel approach in drug delivery. Eur. Polym. J. 2019, 120, 109191.
- Zhang, X.Y.; Wang, Q.; Qin, L.B.; Fu, H.; Fang, Y.W.; Han, B.S.; Duan, Y.R. EGF-modified mPEG-PLGA-PLL nanoparticle for delivering doxorubicin combined with Bcl-2 siRNA as a potential treatment strategy for lung cancer. Drug Deliv. 2016, 23, 2936–2945.
- Yang, D.H.; Kim, H.J.; Park, K.; Kim, J.K.; Chun, H.J. Preparation of poly-L-lysine-based nanoparticles with pH-sensitive release of curcumin for targeted imaging and therapy of liver cancer in vitro and in vivo. Drug Deliv. 2018, 25, 950–960.
- El-Naggar, M.E.; Al-Joufi, F.; Anwar, M.; Attia, M.F.; El-Bana, M.A. Curcumin-loaded PLA-PEG copolymer nanoparticles for treatment of liver inflammation in streptozotocin-induced diabetic rats. Colloid Surf. B 2019, 177, 389–398.
- Cokca, C.; Zartner, L.; Tabujew, I.; Fischer, D.; Peneva, K. Incorporation of Indole Significantly Improves the Transfection Efficiency of Guanidinium-Containing Poly(Methacrylamide)s. Macromol. Rapid. Comm. 2020, 41, 1900668.
- Yu, S.N.; Chen, Y.; Li, X.F.; Gao, Z.L.; Liu, G.F. Chitosan nanoparticle-delivered siRNA reduces CXCR4 expression and sensitizes breast cancer cells to cisplatin. Biosci. Rep. 2017, 37, BSR20170122.
- Nam, J.; Son, S.; Park, K.S.; Moon, J.J. Modularly Programmable Nanoparticle Vaccine Based on Polyethyleneimine for Personalized Cancer Immunotherapy. Adv. Sci. 2021, 8, 2002577.
- Mendonca, M.C.P.; Cronin, M.F.; Cryan, J.F.; O’Driscoll, C.M. Modified cyclodextrin-based nanoparticles mediated delivery of siRNA for huntingtin gene silencing across an in vitro BBB model. Eur. J. Pharm. Biopharm. 2021, 169, 309–318.
- Yang, X.Q.; Lyer, A.K.; Singh, A.; Choy, E.; Hornicek, F.J.; Amiji, M.M.; Duan, Z.F. MDR1 siRNA loaded hyaluronic acid-based CD44 targeted nanoparticle systems circumvent paclitaxel resistance in ovarian cancer. Sci. Rep. 2015, 5, 8509.
- Lu, C.T.; Jin, R.R.; Jiang, Y.N.; Lin, Q.; Yu, W.Z.; Mao, K.L.; Tian, F.R.; Zhao, Y.P.; Zhao, Y.Z. Gelatin nanoparticle-mediated intranasal delivery of substance P protects against 6-hydroxydopamine-induced apoptosis: An in vitro and in vivo study. Drug Des. Dev. Ther. 2015, 9, 1955–1962.
- Lee, S.; Lee, K. pH-Sensitive Folic Acid Conjugated Alginate Nanoparticle for Induction of Cancer-Specific Fluorescence Imaging. Pharmaceutics 2020, 12, 537.
- Huang, G.L.; Huang, H.L. Application of hyaluronic acid as carriers in drug delivery. Drug Deliv. 2018, 25, 766–772.
- Boisguerin, P.; Konate, K.; Josse, E.; Vives, E.; Deshayes, S. Peptide-Based Nanoparticles for Therapeutic Nucleic Acid Delivery. Biomedicines 2021, 9, 583.
- Potzinger, Y.; Rabel, M.; Ahrem, H.; Thamm, J.; Klemm, D.; Fischer, D. Polyelectrolyte layer assembly of bacterial nanocellulose whiskers with plasmid DNA as biocompatible non-viral gene delivery system. Cellulose 2018, 25, 1939–1960.
- Piperno, A.; Sciortino, M.T.; Giusto, E.; Montesi, M.; Panseri, S.; Scala, A. Recent Advances and Challenges in Gene Delivery Mediated by Polyester-Based Nanoparticles. Int. J. Nanomed. 2021, 16, 5981–6002.
- Idrees, H.; Zaidi, S.Z.J.; Sabir, A.; Khan, R.U.; Zhang, X.L.; Hassan, S.U. A Review of Biodegradable Natural Polymer-Based Nanoparticles for Drug Delivery Applications. Nanomaterials 2020, 10, 1970.
- Di Toro, R.; Betti, V.; Spampinato, S. Biocompatibility and integrin-mediated adhesion of human osteoblasts to poly(DL-lactide-co-glycolide) copolymers. Eur. J. Pharm. Sci. 2004, 21, 161–169.
- Cun, D.M.; Jensen, D.K.; Maltesen, M.J.; Bunker, M.; Whiteside, P.; Scurr, D.; Foged, C.; Nielsen, H.M. High loading efficiency and sustained release of siRNA encapsulated in PLGA nanoparticles: Quality by design optimization and characterization. Eur. J. Pharm. Biopharm. 2011, 77, 26–35.
- Dosta, P.; Demos, C.; Ramos, V.; Kang, D.W.; Kumar, S.; Jo, H.; Borros, S. Delivery of siRNA to Endothelial Cells In Vivo Using Lysine/Histidine Oligopeptide-Modified Poly(beta-amino ester) Nanoparticles. Cardiovasc. Eng. Technol. 2021, 12, 114–125.
- Lu, S.; Morris, V.B.; Labhasetwar, V. Effectiveness of Small Interfering RNA Delivery via Arginine-Rich Polyethylenimine-Based Polyplex in Metastatic and Doxorubicin-Resistant Breast Cancer Cells. J. Pharmacol. Exp. Ther. 2019, 370, 902–910.
- Ewe, A.; Hobel, S.; Heine, C.; Merz, L.; Kallendrusch, S.; Bechmann, I.; Merz, F.; Franke, H.; Aigner, A. Optimized polyethylenimine (PEI)-based nanoparticles for siRNA delivery, analyzed in vitro and in an ex vivo tumor tissue slice culture model. Drug Deliv. Transl. Res. 2017, 7, 206–216.
- Du, L.L.; Zhou, J.H.; Meng, L.W.; Wang, X.X.; Wang, C.R.; Huang, Y.Y.; Zheng, S.Q.; Deng, L.D.; Cao, H.Q.; Liang, Z.C.; et al. The pH-Triggered Triblock Nanocarrier Enabled Highly Efficient siRNA Delivery for Cancer Therapy. Theranostics 2017, 7, 3432–3445.
- Zheng, M.; Liu, Y.Y.; Wang, Y.B.; Zhang, D.Y.; Zou, Y.; Ruan, W.M.; Yin, J.L.; Tao, W.; Park, J.B.; Shi, B.Y. ROS-Responsive Polymeric siRNA Nanomedicine Stabilized by Triple Interactions for the Robust Glioblastoma Combinational RNAi Therapy. Adv. Mater. 2019, 31, e1903277.
This entry is offline, you can click here to edit this entry!
