There is an urgent and imminent need to develop new antimicrobials to fight against antibiotic-resistant bacterial and fungal strains. In this study, a checkerboard method was used to evaluate the synergistic effects of the antimicrobial peptide P-113 and its bulky non-nature amino acid substituted derivatives with vancomycin against vancomycin-resistant Enterococcus faecium, Staphylococcus aureus, and wild-type Escherichia coli. Boron-dipyrro-methene (BODIPY) labeled vancomycin was used to characterize the interactions between the peptides, vancomycin, and bacterial strains. Moreover, neutralization of antibiotic-induced releasing of lipopolysaccharide (LPS) from E. coli by the peptides was obtained. Among these peptides, Bip-P-113 demonstrated the best minimal inhibitory concentrations (MICs), antibiotics synergism, bacterial membrane permeabilization, and supernatant LPS neutralizing activities against the bacteria studied. These results could help in developing antimicrobial peptides that have synergistic activity with large size glycopeptides such as vancomycin in therapeutic applications.
- antimicrobial peptide
- antibiotic resistance
- vancomycin
- synergism
- bulky non-nature amino acid
1. Introduction
Previously, We have developed a method to increase salt resistance, serum proteolytic stability, and LPS neutralizing activities of antimicrobial peptides by adding bulky non-nature amino acids to their termini [1][2]. In this study, we want to investigate the antibacterial activity of P-113 and its derivatives (Table 1) and to assess their ability to be used in combination with vancomycin against E. coli as well as vancomycin-resistant Gram-positive bacteria. The molecular mechanisms of the synergistic effect between P-113 and its derivatives and vancomycin were also studied.
Table 1. Sequences of P-113, Phe-P-113, Bip-P-113, Dip-P-113 and Nal-P-113.
| Name | Chemical Structure a | Sequence b | Molecular Weight (Da) |
|---|---|---|---|
| P-113 | 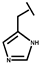 |
Ac- A K R His His G Y K R K F His -NH2 | 1605.86 |
| Phe-P-113 | 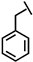 |
Ac- A K R Phe Phe G Y K R K F Phe -NH2 | 1635.99 |
| Bip-P-113 | 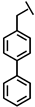 |
Ac- A K R Bip Bip G Y K R K F Bip -NH2 | 1864.97 |
| Dip-P-113 | 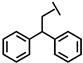 |
Ac- A K R Dip Dip G Y K R K F Dip -NH2 | 1864.06 |
| Nal-P-113 | 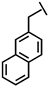 |
Ac- A K R Nal Nal G Y K R K F Nal -NH2 | 1786.03 |
a Side chain structure of amino acids at positions 4, 5, 12. b Bip: β-(4.4’-biphenyl)alanine; Dip: β-diphenylalanine; Nal: β-Naphthylalanine.
2. Synergistic Effect with Vancomycin in the Presence of a Sub-Inhibitory Concentration (¼ ×MIC) of Peptides
Due to the strong antimicrobial activity, bacterial membrane permeability, and killing kinetics, we hypothesized that these peptides could be used to resensitize vancomycin-resistant Gram-positive bacterial strains against vancomycin and even to have antibacterial activities against Gram-negative bacterial strains such as E. coli. To assess this goal, synergetic activities of P-113 and its derivatives at a sub-inhibitory concentration (¼ ×MIC) with vancomycin were determined by the checkerboard assay as described previously (Table 2). The fractional inhibitory concentration indices (FICI) were interpreted as follows: ≤ 0.5, “synergy”; > 0.5–4, “no interaction”; and > 4, “antagonism” . Both P-113 and Phe-P-113 showed no synergy while combined with vancomycin. Dip-P-113 had only synergistic effects against VRE, VRSA 02, and E. coli. Nal-P-113 had synergistic effects against VRE, VISA 03, VRSA 01, VRSA 02, VRSA 03, and E. coli. Bip-P-113 demonstrated the best synergy with vancomycin against all of the bacterial strains tested.
Table 3. Synergistic effects of P-113, Phe-P-113, Bip-P-113, Dip-P-113, and Nal-P-113 in combination with vancomycin against bacterial strains studied. All of the experiments were performed in triplicate.
| Strains | AMP (μg/mL) (¼ ×MIC) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P-113 | Phe-P-113 | Bip-P-113 | Dip-P-113 | Nal-P-113 | ||||||
| VAN a | FICI b | VAN | FICI | VAN | FICI | VAN | FICI | VAN | FICI | |
| VRE | 32 | 0.75 | 32 | 0.75 | 8 | 0.38 | 16 | 0.50 | 16 | 0.50 |
| VISA 01 | 4 | 1.25 | 4 | 1.25 | 1 | 0.50 | 4 | 1.25 | 2 | 0.75 |
| VISA 02 | 4 | 1.25 | 4 | 1.25 | 1 | 0.50 | 4 | 1.25 | 2 | 0.75 |
| VISA 03 | 4 | 1.25 | 4 | 1.25 | 1 | 0.50 | 2 | 0.75 | 1 | 0.50 |
| VRSA 01 | 32 | 1.25 | 32 | 1.25 | 2 | 0.31 | 16 | 0.75 | 8 | 0.50 |
| VRSA 02 | 32 | 1.25 | 32 | 1.25 | 2 | 0.31 | 8 | 0.50 | 8 | 0.50 |
| VRSA 03 | 32 | 1.25 | 32 | 1.25 | 4 | 0.38 | 16 | 0.75 | 4 | 0.38 |
| E. coli ATCC 25922 |
>64 | 1.25 | 64 | 0.75 | 16 | 0.38 | 32 | 0.50 | 16 | 0.38 |
a VAN, vancomycin (μg/mL). b FICI, fractional inhibitory concentration index, FICI ≤ 0.5, synergy; 0.5 < FICI ≤ 4, no interaction; FICI > 4, antagonism [3].
3. Mechanism of Resensitization of Bacteria to Vancomycin by P-113 and its Derivatives
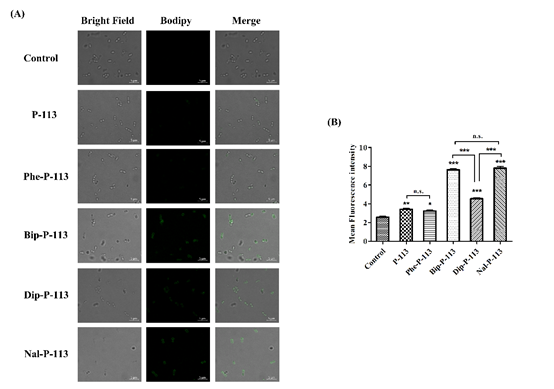
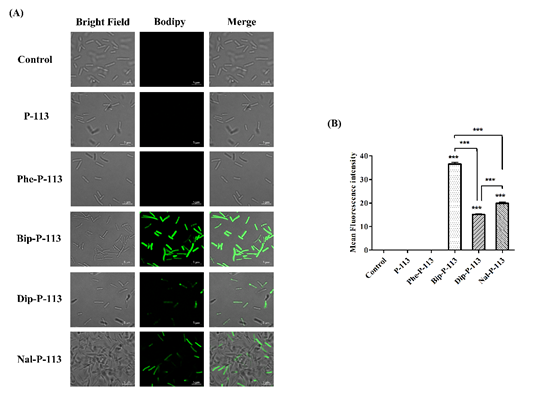
We then used boron-dipyrro-methene (BODIPY) labeled vancomycin to study the possible mechanism of synergism between the designed peptides, vancomycin, and the bacterial strains. The results demonstrated that P-113 derivatives with bulky non-nature amino acids markedly enhanced the entry of BODIPY-labeled vancomycin into the VRE strain studied with the order Bip-P-113 = Nal-P-113 > Dip-P-113 > Phe-P-113 = P-113 (Figure 1). Similarly, the order of the BODIPY-labeled vancomycin entering into the Gram-negative E. coli strain is Bip-P-113 > Nal-P-113 > Dip-P-113 > Phe-P-113 = P-113 (Figure 2).
Figure 1. P-113 and its derivatives increase the uptake of BODIPY-labeled vancomycin in E. faecium. (A) Fluorescence images of 107 CFU/mL E. faecium BCRC 15B0132 treated with 0.5 ×MIC of P-113, Phe-P-113, Bip-P-113, Dip-P-113, and Nal-P-113 at 37 °C for 30 min, then treated BODIPY-labeled vancomycin for 30 min (scale bar represents 5 µm). (B) Mean fluorescence intensity of P-113, Phe-P-113, Bip-P-113, Dip-P-113, and Nal-P-113 treated in E. faecium BCRC 15B0132. Samples treated with BODIPY-labeled vancomycin only served as a control. Results are presented as means ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001 compared with control. n.s. = no significant differences. The experiments were performed in triplicate.
Figure 2. P-113 and its derivatives increase the uptake of BODIPY-labeled vancomycin in E. coli. (A) Fluorescence images of 107 CFU/mL E. coli ATCC 25922 treated with 0.5 ×MIC of P-113, Phe-P-113, Bip-P-113, Dip-P-113, and Nal-P-113 at 37 °C for 30 min, then treated with BODIPY-labeled vancomycin for 30 min (scale bar represents 5 µm). (B) Mean fluorescence intensity of P-113, Phe-P-113, Bip-P-113, Dip-P-113, and Nal-P-113 treated in E. coli ATCC 25922. Samples treated with BODIPY-labeled vancomycin only served as a control. Results are presented as means ± SEM, *** p < 0.001 compared with control. The experiments were performed in triplicate.
4. P-113 and its Derivatives Attenuate Vancomycin-Induced LPS Release
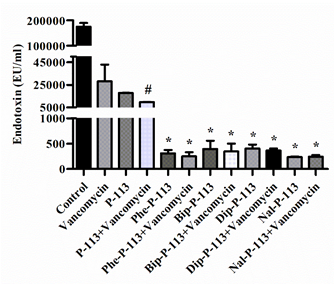
In the present study, we measured the concentration of LPS in supernatants and investigated the effect of P-113 and its derivatives on vancomycin-induced release of LPS by limulus amebocyte lysate (LAL) assay (Figure 3). When E. coli cells were treated with the bulky non-nature amino acids substituted P-113 derivatives or the combination of vancomycin and P-113 derivatives, the concentration of LPS in the supernatants decreased tremendously compared to the PBS-treated control cultures (Figure 3).
Figure 3. The influence of vancomycin, P-113 and its derivatives on LPS release from E. coli. E. coli ATCC 25922 was incubated at the mid-log phase (104 CFU/mL) and treated with peptide alone (at 1 × MIC) or combination with vancomycin (both at 0.5 × MIC) at 37 °C for 6 h. The samples were filtered through 0.2 µm pore filter and the endotoxin level was detected by LAL assay. # p < 0.05 compared with vancomycin only, * p < 0.001 compared with vancomycin only. The experiments were performed in triplicate.
5. Conclusions
In conclusion, our results demonstrated that the bulky non-nature amino acid substituted P-113 derivatives display compelling antibacterial and synergistic activities with vancomycin against vancomycin-resistant E. faecium, S. aureus, and wild-type E. coli. Moreover, these antimicrobial peptides also reduced antibiotic-induced releasing of LPS from Gram-negative bacteria. Among these peptides, Bip-P-113 demonstrated the best MICs, antibiotics synergism, and supernatant LPS neutralizing activities against the bacteria studied. Our results should be useful in the development of new antimicrobial peptides and peptidomimetics with both antimicrobial activity and antibiotics synergism for potential therapeutic applications.
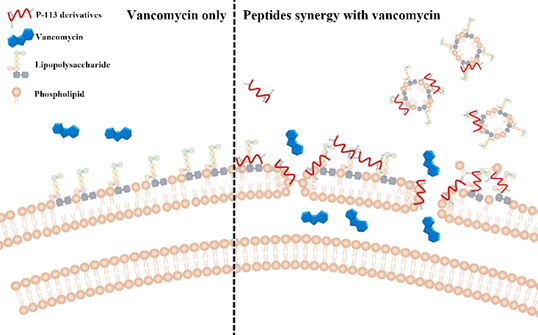
Figure 4. Synergistic mechanism of action proposed for P-113 derivatives and vancomycin. For vancomycin, the efficacy was limited due to the permeability barrier of the outer membrane of Gram-negative bacteria (left panel). However, P-113 derivatives (e.g., Bip-P-113, Dip-P-113, and Nal-P-113) could disturb the outer membrane of Gram-negative bacteria and enhance the entry of vancomycin into these bacteria (right panel). Moreover, P-113 derivatives could bind and neutralize LPS through extra hydrophobic interactions with the lipid A motif of LPS (right panel).
References
- Chih, Y.H.; Lin, Y.S.; Yip, B.S.; Wei, H.J.; Chu, H.L.; Yu, H.Y.; Cheng, H.T.; Chou, Y.T.; Cheng, J.W. Ultrashort antimicrobial peptides with antiendotoxin properties. Agents Chemother. 2015, 59, 5052–5056.
- Chu, H.L.; Yu, H.Y.; Yip, B.S.; Chih, Y.H.; Liang, C.W.; Cheng, H.T.; Cheng, J.W. Boosting salt resistance of short antimicrobial peptides. Agents Chemother. 2013, 57, 4050–4052.
- Feng, Q.; Huang, Y.; Chen, M.; Li, G.; Chen, Y. Functional synergy of a-helical antimicrobial peptides and traditional antibiotics against Gram-negative and Gram-positive bacteria in vitro and in vivo. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 197–204.