Immune response has been shown to play an important role in defining patient prognosis and response to cancer treatment. Tumor-induced immunosuppression encouraged the recent development of new chemotherapeutic agents that assists in the augmentation of immune responses. Molecular mechanisms that tumors use to evade immunosurveillance are attributed to their ability to alter antigen processing/presentation pathways and the tumor microenvironment. Cancer cells take advantage of normal molecular and immunoregulatory machinery to survive and thrive. Cancer cells constantly adjust their genetic makeup using several mechanisms such as nucleotide excision repair as well as microsatellite and chromosomal instability, thus giving rise to new variants with reduced immunogenicity and the ability to continue to grow without restrictions.
- immunosuppression
- Cancer
- Signaling Pathways
1. Introduction
2. Immune Evasion
2.1. Elimination
2.2. Equilibrium
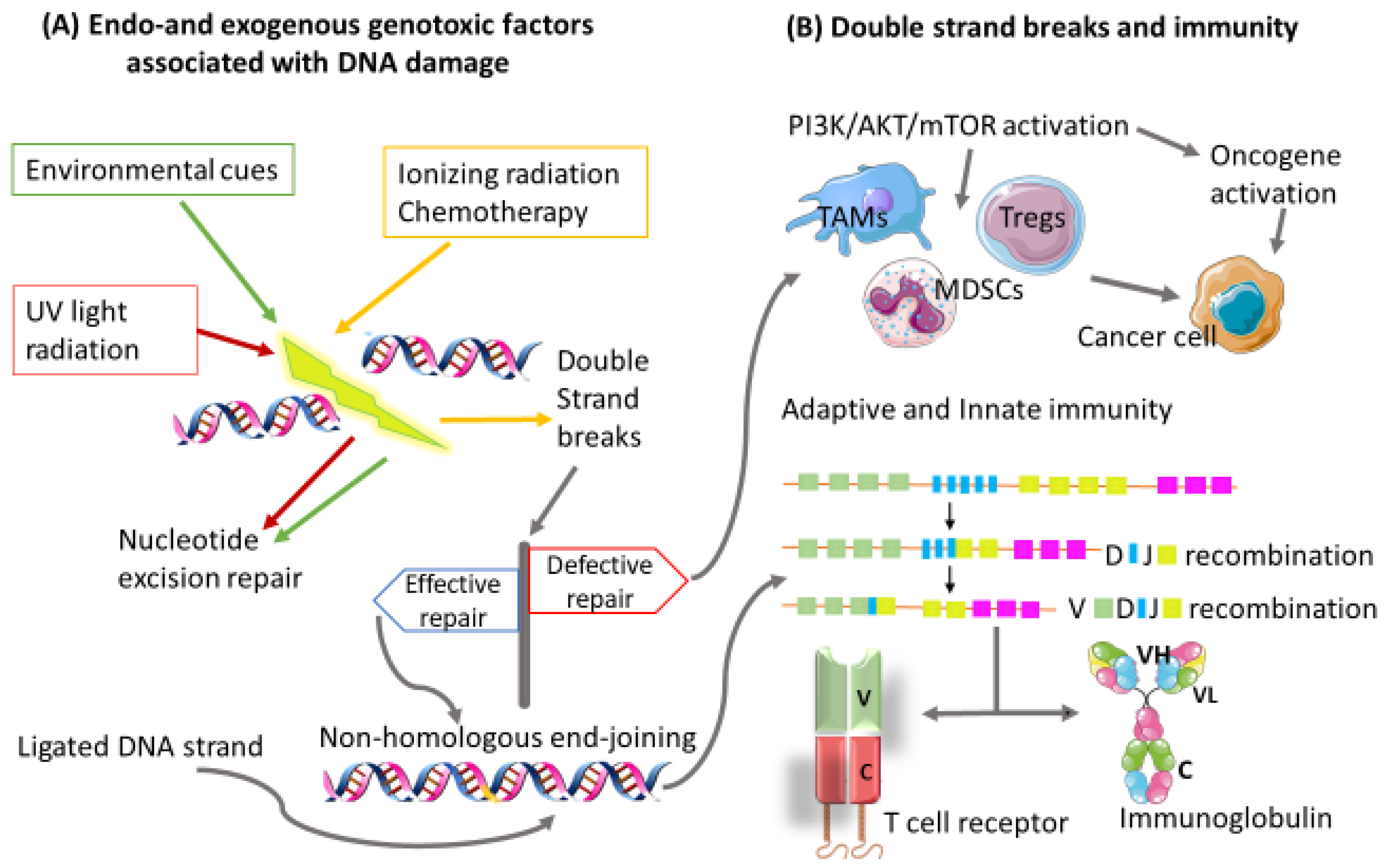
2.3. Escape
2.3.1. Escaping the Antigen Presentation Pathway
2.3.2. The PD-1/PD-L1 Pathway as a Mechanism of Escape
3. The Tumor Microenvironment
4. Immunoregulatory Signaling Pathways
4.1. Myeloid-Derived Suppressor Cells
4.2. Tumor-Associated Macrophages
4.3. Regulatory T Cells
5. Personalized Precision Medicine and Combinatorial Therapies
6. Conclusions and Future Perspectives
| PI3K Inhibitor | Mode of Action | Cancer Type | References |
|---|---|---|---|
| Alpelisib | PIK3CA/PI3K-δ isoform | Hormone receptor +/HER2-Breast Cancer | [52][53] |
| Copanlisib | PI3Kβ, PI3Kγ, PI3K-α & PI3K-δ isoforms | Follicular Lymphoma | [54] |
| Duvelisib | PI3K-δ and PI3K γ isoforms | Chronic Lymphocytic Leukemia | [55] |
| Idelalisib | PI3Kδ | Chronic Lymphocytic Leukemia | [56] |
| Buparlisib | PI3Kα, PI3Kβ, PI3Kδ and PI3Kγ | Metastatic triple-negative breast and colorectal cancers | [51][57] |
| Pictilisib | PI3Kα and PI3Kδ | Advanced breast cancer and cancer of the bone | [58][59] |
This entry is adapted from the peer-reviewed paper 10.3390/biomedicines10030682
References
- Li Kai, Luo Haiqing, Huang Lianfang, Luo Hui, Zhu Xiao; Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int 2020, 20, 16, 10.1186/s12935-019-1091-8. .
- do Canto, L. M; Larsen, S. J; Catin Kupper, B. E; Begnami, M; Scapulatempo-Neto, C; Petersen, A. H; Aagaard, M. M; Baumbach, J; Aguiar, S Jr; & Rogatto, S. R.; et al. Increased Levels of Genomic Instability and Mutations in Homologous Recombination Genes in Locally Advanced Rectal Carcinomas. Frontiers in oncology 2019, 9, 395, 10.3389/fonc.2019.00395.
- Giwon Shin; Stephanie U. Greer; Erik Hopmans; Susan M. Grimes; Hojoon Lee; Lan Zhao; Laura Miotke; Carlos Suarez; Alison F. Almeda; Sigurdis Haraldsdottir; et al. Profiling diverse sequence tandem repeats in colorectal cancer reveals co-occurrence of microsatellite and chromosomal instability involving Chromosome 8. Genome Medicine 2021, 13, 1-18, 10.1186/s13073-021-00958-z.
- Jardim J. Melanie; Wang Qinhong; Furumai Ryohei; Wakeman Timothy; Goodman K. Barbara; Wang Xiao-Fan; Reduced ATR or Chk1 Expression Leads to Chromosome Instability and Chemosensitization of Mismatch Repair–deficient Colorectal Cancer Cells. Mol. Biol. Cell 2009, 20, 3801–3809, doi.org/10.1091/mbc.e09-04-0303.
- Chen Mengting; Renske Linstra; van Vugt A.T.M. Marcel; Genomic instability, inflammatory signaling and response to cancer immunotherapy. Biochim. et Biophys. Acta (BBA) Rev. Cancer 2022, 1877, 188661, https://doi.org/10.1016/j.bbcan.2021.188661.
- Ryungsa Kim; Manabu Emi; Kazuaki Tanabe; Koji Arihiro; Tumor-Driven Evolution of Immunosuppressive Networks during Malignant Progression. Cancer Research 2006, 66, 5527-5536, 10.1158/0008-5472.can-05-4128.
- DaQian Gu; Xiang Ao; Yu Yang; Zhuo Chen; Xiang Xu; Soluble immune checkpoints in cancer: production, function and biological significance. Journal for ImmunoTherapy of Cancer 2018, 6, 132, 10.1186/s40425-018-0449-0.
- Rebekka Weber; Viktor Fleming; Xiaoying Hu; Vasyl Nagibin; Christopher Groth; Peter Altevogt; Jochen Utikal; Viktor Umansky; Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Frontiers in Immunology 2018, 9, 1310, 10.3389/fimmu.2018.01310.
- Ruth Nussinov; Chung-Jung Tsai; Hyunbum Jang; A New View of Pathway-Driven Drug Resistance in Tumor Proliferation. Trends in Pharmacological Sciences 2017, 38, 427-437, 10.1016/j.tips.2017.02.001.
- Ehrlich, P; Ueber den jetzigen stand der karzinomforschung. vortrag gehalten vor den studenten der amsterdamer universitaet, vereinigung fuer wissenschaftliche arbeit 1 june 1908.. Beitraege Exp. Pathol. Chemother. Akad. Verl. Leipz 1909, 118, 164, .
- MacFarlane Burnet; Cancer--A Biological Approach: III. Viruses Associated with Neoplastic Conditions. IV. Practical Applications. BMJ 1957, 1, 841-847, 10.1136/bmj.1.5023.841.
- L Thomas; On immunosurveillance in human cancer.. The Yale journal of biology and medicine 1982, 55, 329-33, .
- Gavin P. Dunn; Lloyd J. Old; Robert D. Schreiber; The Immunobiology of Cancer Immunosurveillance and Immunoediting. Immunity 2004, 21, 137-148, 10.1016/j.immuni.2004.07.017.
- Joanina K. Gicobi; Whitney Barham; Haidong Dong; Immune resilience in response to cancer therapy. Cancer Immunology, Immunotherapy 2020, 69, 2165-2167, 10.1007/s00262-020-02731-4.
- Adriana Albini; Antonino Bruno; Douglas M. Noonan; Lorenzo Mortara; Contribution to Tumor Angiogenesis From Innate Immune Cells Within the Tumor Microenvironment: Implications for Immunotherapy. Frontiers in Immunology 2018, 9, 527, 10.3389/fimmu.2018.00527.
- Sergei I. Grivennikov; Florian Greten; Michael Karin; Immunity, Inflammation, and Cancer. Cell 2010, 140, 883-899, 10.1016/j.cell.2010.01.025.
- Mark G Goldstein; Zihai Li; Heat-shock proteins in infection-mediated inflammation-induced tumorigenesis. Journal of Hematology & Oncology 2009, 2, 5-5, 10.1186/1756-8722-2-5.
- I. Seren Bernardone; Role of NK cells and adaptive immunity in “immunoediting”: Recent developments. Inmunología 2008, 27, 141-146, 10.1016/s0213-9626(08)70062-3.
- Jing Zhang; David J. H. Shih; Shiaw-Yih Lin; Role of DNA repair defects in predicting immunotherapy response. Biomarker Research 2020, 8, 1-8, 10.1186/s40364-020-00202-7.
- Myrthe Jager; Francis Blokzijl; Ewart Kuijk; Johanna Bertl; Maria Vougioukalaki; Roel Janssen; Nicolle Besselink; Sander Boymans; Joep de Ligt; Jakob Skou Pedersen; et al. Deficiency of nucleotide excision repair is associated with mutational signature observed in cancer. Genome Research 2019, 29, 1067-1077, 10.1101/gr.246223.118.
- Natalia Vargas-Rondón; Victoria E. Villegas; Milena Rondón-Lagos; The Role of Chromosomal Instability in Cancer and Therapeutic Responses. Cancers 2017, 10, 4, 10.3390/cancers10010004.
- Ryungsa Kim; Manabu Emi; Kazuaki Tanabe; Cancer immunoediting from immune surveillance to immune escape. Immunology 2007, 121, 1-14, 10.1111/j.1365-2567.2007.02587.x.
- Lynnette R. Ferguson; Helen Chen; Andrew R. Collins; Marisa Connell; Giovanna Damia; Santanu Dasgupta; Meenakshi Malhotra; Alan K. Meeker; Amedeo Amedei; Amr Amin; et al. Genomic instability in human cancer: Molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Seminars in Cancer Biology 2015, 35, S5-S24, 10.1016/j.semcancer.2015.03.005.
- Yixin Yao Wei Dai; Genomic Instability and Cancer. Journal of Carcinogenesis & Mutagenesis 2014, 5, 1000165, 10.4172/2157-2518.1000165.
- Yixin Yao Wei Dai; Genomic Instability and Cancer. Journal of Carcinogenesis & Mutagenesis 2014, 5, 1000165, 10.4172/2157-2518.1000165.
- Daan K. J. Pieren; Noortje A. M. Smits; Sandra Imholz; Bhawani Nagarajah; Conny T. van Oostrom; Renata M. C. Brandt; Wilbert P. Vermeij; Martijn E. T. Dollé; Teun Guichelaar; Compromised DNA Repair Promotes the Accumulation of Regulatory T Cells With an Aging-Related Phenotype and Responsiveness. Frontiers in Aging 2021, 2, 13, 10.3389/fragi.2021.667193.
- Laetitia Nebot-Bral; Clélia Coutzac; Patricia L. Kannouche; Nathalie Chaput; Why is immunotherapy effective (or not) in patients with MSI/MMRD tumors?. Bulletin du Cancer 2018, 106, 105-113, 10.1016/j.bulcan.2018.08.007.
- Salihanur Darici; Hazem AlKhaldi; Gillian Horne; Heather G. Jørgensen; Sandra Marmiroli; Xu Huang; Targeting PI3K/Akt/mTOR in AML: Rationale and Clinical Evidence. Journal of Clinical Medicine 2020, 9, 2934, 10.3390/jcm9092934.
- Zhilin Zou; Tao Tao; Hongmei Li; Xiao Zhu; mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell & Bioscience 2020, 10, 1-11, 10.1186/s13578-020-00396-1.
- Luca Cassetta; Jeffrey W. Pollard; Tumor-associated macrophages. Current Biology 2020, 30, R246-R248, 10.1016/j.cub.2020.01.031.
- Amanda Valeta-Magara; Abhilash Gadi; Viviana Volta; Beth Walters; Rezina Arju; Shah Giashuddin; Hua Zhong; Robert J. Schneider; Inflammatory Breast Cancer Promotes Development of M2 Tumor-Associated Macrophages and Cancer Mesenchymal Cells through a Complex Chemokine Network. Cancer Research 2019, 79, 3360-3371, 10.1158/0008-5472.can-17-2158.
- Yahui Zhao; Weina Zhang; Miaomiao Huo; Peng Wang; Xianghe Liu; Yu Wang; Yinuo Li; Zhixiang Zhou; Ningzhi Xu; Hongxia Zhu; et al. XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer. Signal Transduction and Targeted Therapy 2021, 6, 1-14, 10.1038/s41392-021-00761-7.
- Samantha B Kemp; Nina G Steele; Eileen S Carpenter; Katelyn L Donahue; Grace G Bushnell; Aaron H Morris; Stephanie The; Sophia M Orbach; Veerin R Sirihorachai; Zeribe C Nwosu; et al. Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Science Alliance 2021, 4, e202000935, 10.26508/lsa.202000935.
- Prahara Yuri; Katsumi Shigemura; Koichi Kitagawa; Exsa Hadibrata; Muhammad Risan; Andy Zulfiqqar; Indrawarman Soeroharjo; Ahmad Z. Hendri; Raden Danarto; Aya Ishii; et al. Increased tumor-associated macrophages in the prostate cancer microenvironment predicted patients’ survival and responses to androgen deprivation therapies in Indonesian patients cohort. Prostate International 2020, 8, 62-69, 10.1016/j.prnil.2019.12.001.
- S. A. Almatroodi; C. F. McDonald; I. A. Darby; D. S. Pouniotis; Characterization of M1/M2 Tumour-Associated Macrophages (TAMs) and Th1/Th2 Cytokine Profiles in Patients with NSCLC. Cancer Microenvironment 2015, 9, 1-11, 10.1007/s12307-015-0174-x.
- Antonio Sica; Vincenzo Bronte; Altered macrophage differentiation and immune dysfunction in tumor development. Journal of Clinical Investigation 2007, 117, 1155-1166, 10.1172/jci31422.
- Antonio Sica; Alberto Mantovani; Macrophage plasticity and polarization: in vivo veritas. Journal of Clinical Investigation 2012, 122, 787-795, 10.1172/jci59643.
- Tao Yu; Shucheng Gan; Qingchen Zhu; Dongfang Dai; Ni Li; Hui Wang; Xiaosong Chen; Dan Hou; Yan Wang; Qiang Pan; et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nature Communications 2019, 10, 1-15, 10.1038/s41467-019-12384-2.
- Cuiping Fu; Liyan Jiang; Shengyu Hao; Zilong Liu; Suling Ding; Weiwei Zhang; Xiangdong Yang; Shanqun Li; Activation of the IL-4/STAT6 Signaling Pathway Promotes Lung Cancer Progression by Increasing M2 Myeloid Cells. Frontiers in Immunology 2019, 10, 2638, 10.3389/fimmu.2019.02638.
- Nan Wang; Hongwei Liang; Ke Zen; Molecular Mechanisms That Influence the Macrophage M1–M2 Polarization Balance. Frontiers in Immunology 2014, 5, 614, 10.3389/fimmu.2014.00614.
- Shiyang Yuan; Yaling Dong; Laishui Peng; Mei Yang; Linxia Niu; Zhiwen Liu; Junping Xie; Tumor‑associated macrophages affect the biological behavior of lung adenocarcinoma A549 cells through the PI3K/AKT signaling pathway. Oncology Letters 2019, 18, 1840-1846, 10.3892/ol.2019.10483.
- Mariane T. Amano; Angela Castoldi; Vinicius Andrade-Oliveira; Marcela T. Latancia; Fernanda F. Terra; Matheus Correa-Costa; Cristiane N.S. Breda; Raphael J.F. Felizardo; Welbert O. Pereira; Marina B. da Silva; et al. The lack of PI3Kγ favors M1 macrophage polarization and does not prevent kidney diseases progression. International Immunopharmacology 2018, 64, 151-161, 10.1016/j.intimp.2018.08.020.
- Chunmei Liu; Bohui Li; Kaihong Tang; Xuening Dong; Longge Xue; Guangming Su; Yingyu Jin; Aquaporin 1 alleviates acute kidney injury via PI3K-mediated macrophage M2 polarization. Inflammation Research 2020, 69, 509-521, 10.1007/s00011-020-01334-0.
- Marina Pasca di Magliano; Matthias Hebrok; Hedgehog signalling in cancer formation and maintenance. Nature Reviews Cancer 2003, 3, 903-911, 10.1038/nrc1229.
- Amy J. Petty; Ang Li; Xinyi Wang; Rui Dai; Benjamin Heyman; David Hsu; XiaoPei Huang; Yiping Yang; Hedgehog signaling promotes tumor-associated macrophage polarization to suppress intratumoral CD8+ T cell recruitment. Journal of Clinical Investigation 2019, 129, 5151-5162, 10.1172/jci128644.
- Jing Yang; Ji Nie; Xuelei Ma; Yuquan Wei; Yong Peng; Xiawei Wei; Targeting PI3K in cancer: mechanisms and advances in clinical trials. Molecular Cancer 2019, 18, 1-28, 10.1186/s12943-019-0954-x.
- Ying Shao; William Y. Yang; Fatma Saaoud; Charles Drummer; Yu Sun; Keman Xu; Yifan Lu; Huimin Shan; Ethan M. Shevach; Xiaohua Jiang; et al. IL-35 promotes CD4+Foxp3+ Tregs and inhibits atherosclerosis via maintaining CCR5-amplified Treg-suppressive mechanisms. JCI Insight 2021, 6, e152511, 10.1172/jci.insight.152511.
- Ali S. Alzahrani; PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Seminars in Cancer Biology 2019, 59, 125-132, 10.1016/j.semcancer.2019.07.009.
- Nakamura, R.M.; Kasahara, Y. Chapter 19—Molecular Diagnostics in the Evaluation of Cancer: Modern Concepts and Overview; Grody, W.W.; Nakamura, R.M.; Strom, C.M.; Kiechle, F.L, Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 215–223.
- Jennifer H. Gunter; Marianna Kruithof-De Julio; Eugenio Zoni; Editorial: Personalized Medicine for Urological Cancers: Targeting Cancer Metabolism. Frontiers in Oncology 2022, 12, 862811, 10.3389/fonc.2022.862811.
- Ana C. Garrido-Castro; Cristina Saura; Romualdo Barroso-Sousa; Hao Guo; Eva Ciruelos; Begoña Bermejo; Joaquin Gavilá; Violeta Serra; Aleix Prat; Laia Paré; et al. Phase 2 study of buparlisib (BKM120), a pan-class I PI3K inhibitor, in patients with metastatic triple-negative breast cancer. Breast Cancer Research 2020, 22, 1-13, 10.1186/s13058-020-01354-y.
- Tori Wilhoit; PharmD Jeannie M. Patrick; PharmD Megan B. May; Alpelisib: A Novel Therapy for Patients With PIK3CA-Mutated Metastatic Breast Cancer. Journal of the Advanced Practitioner in Oncology 2020, 11, 768-775, 10.6004/jadpro.2020.11.7.9.
- B. Verret; J. Cortes; T. Bachelot; F. Andre; Monica Arnedos; Efficacy of PI3K inhibitors in advanced breast cancer. Annals of Oncology 2019, 30, x12-x20, 10.1093/annonc/mdz381.
- Felix A Mensah; Jean-Pierre Blaize; Locke J Bryan; Spotlight on copanlisib and its potential in the treatment of relapsed/refractory follicular lymphoma: evidence to date. OncoTargets and Therapy 2018, ume 11, 4817-4827, 10.2147/ott.s142264.
- Hima V. Vangapandu; Nitin Jain; Varsha Gandhi; Duvelisib: a phosphoinositide-3 kinase δ/γ inhibitor for chronic lymphocytic leukemia. Expert Opinion on Investigational Drugs 2017, 26, 625-632, 10.1080/13543784.2017.1312338.
- Chan Yoon Cheah; Nathan H. Fowler; Idelalisib in the management of lymphoma. Blood 2016, 128, 331-336, 10.1182/blood-2016-02-702761.
- Jordi Rodon; Irene Brana; Lillian L Siu; Maja J De Jonge; Natasha Homji; David Mills; Emmanuelle Di Tomaso; Celine Sarr; Lucia Trandafir; Cristian Massacesi; et al. Phase I dose-escalation and -expansion study of buparlisib (BKM120), an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. Investigational New Drugs 2014, 32, 670-681, 10.1007/s10637-014-0082-9.
- Ian E Krop; Ingrid A Mayer; Vinod Ganju; Maura Dickler; Stephen Johnston; Serafin Morales; Denise A Yardley; Bohuslav Melichar; Andres Forero-Torres; Soo Chin Lee; et al. Pictilisib for oestrogen receptor-positive, aromatase inhibitor-resistant, advanced or metastatic breast cancer (FERGI): a randomised, double-blind, placebo-controlled, phase 2 trial. The Lancet Oncology 2016, 17, 811-821, 10.1016/s1470-2045(16)00106-6.
- Chao Liang; Xijiao Yu; Naping Xiong; Zhichang Zhang; Zhenyu Sun; Yang Dong; Pictilisib Enhances the Antitumor Effect of Doxorubicin and Prevents Tumor-Mediated Bone Destruction by Blockade of PI3K/AKT Pathway. Frontiers in Oncology 2021, 10, 615146, 10.3389/fonc.2020.615146.
