Neuroimaging is a widely available, less-invasive method to investigate the whole brain of humans, and with neuroimaging, the brain’s morphological and microstructural features can be obtained. A neuroimaging-based brain-age estimation can provide a reliable neuropsychiatric biomarker at the single-subject level. In addition, brain MRI—particularly T1-weighted structural Magnetic resonance imaging (MRI)—is a widely available examination in most countries, which may support easier and wider clinical applications of brain-age analyses. Considering the utility, availability, and reproducibility of neuroimaging-based brain-age estimations for single patients, brain age can be expected to become a useful personalized biomarker in neuropsychiatry.
- brain age
- neuropsychiatric disorder
- neuroimaging
- machine learning
1. Aging, Disease, and the Brain
2. Neuroimaging-Based Brain Age Estimation
2.1. Theory of Neuroimaging-Based Brain-Age Estimation
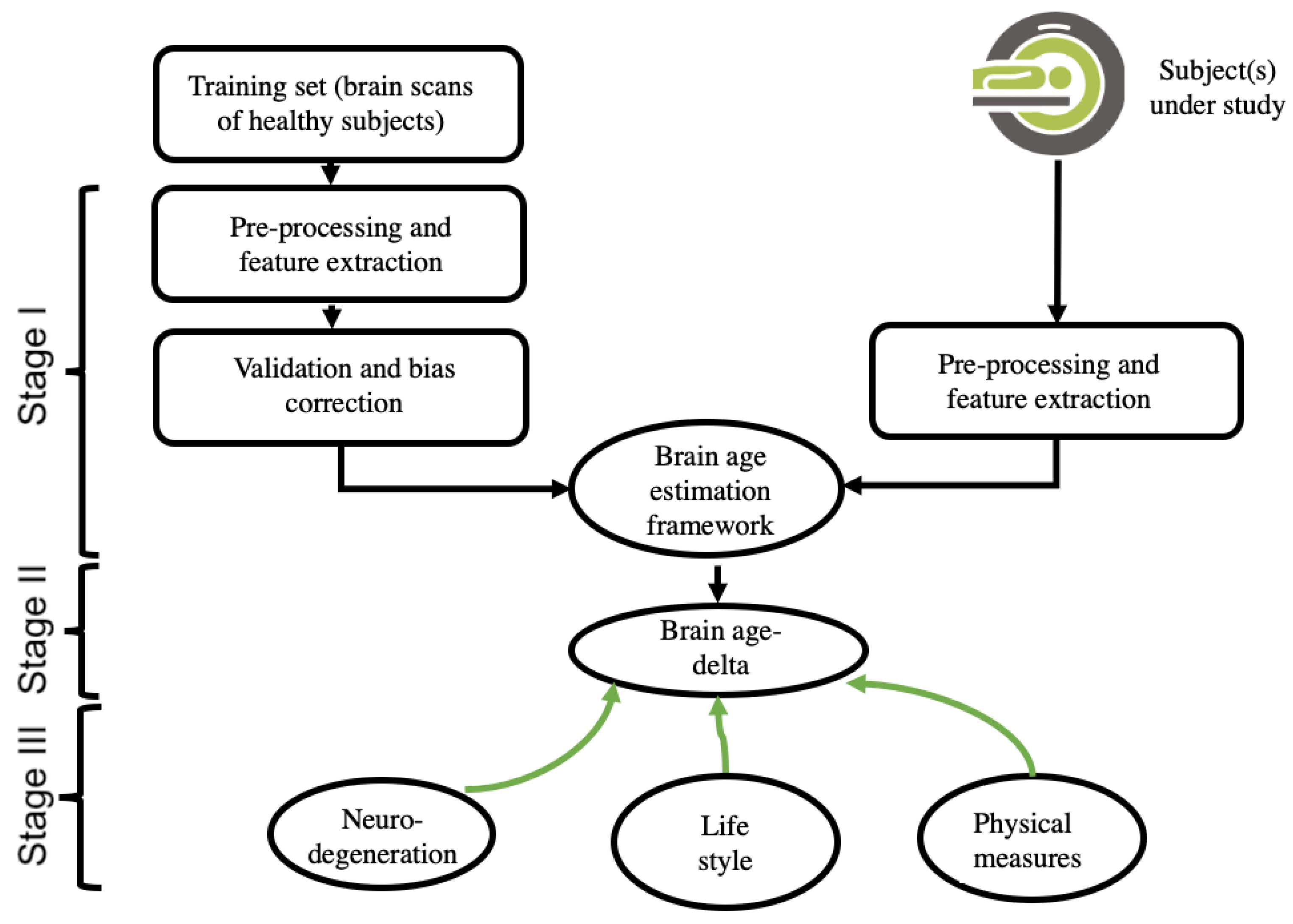
2.2. Input Data and Feature-Extraction Methodologies of Neuroimaging
2.3. Data Reduction, Validation, and Bias Adjustment Neuroimaging Methodologies
2.4. Machine-Learning Methodologies
This entry is adapted from the peer-reviewed paper 10.3390/jpm12111850
References
- Flatt, T. A New Definition of Aging? Front. Genet. 2012, 3, 148.
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198.
- Childs, B.G.; Durik, M.; Baker, D.J.; Van Deursen, J.M. Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat. Med. 2015, 21, 1424–1435.
- Franke, K.; Bublak, P.; Hoyer, D.; Billiet, T.; Gaser, C.; Witte, O.; Schwab, M. In vivo biomarkers of structural and functional brain development and aging in humans. Neurosci. Biobehav. Rev. 2020, 117, 142–164.
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581.
- Azam, S.; Haque, E.; Balakrishnan, R.; Kim, I.-S.; Choi, D.-K. The Ageing Brain: Molecular and Cellular Basis of Neurodegeneration. Front. Cell Dev. Biol. 2021, 9, 683459.
- Hochla, N.A.N.; Fabian, M.S.; Parsons, O.A. Brain-age quotients in recently detoxified alcoholic, recovered alcoholic and nonalcoholic women. J. Clin. Psychol. 1982, 38, 207–212.
- Franke, K.; Gaser, C. Ten Years of BrainAGE as a Neuroimaging Biomarker of Brain Aging: What Insights Have We Gained? Front. Neurol. 2019, 10, 789.
- Franke, K.; Ziegler, G.; Klöppel, S.; Gaser, C.; Alzheimer’s Disease Neuroimaging Initiative. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. NeuroImage 2010, 50, 883–892.
- Cole, J.H. Multimodality neuroimaging brain-age in UK biobank: Relationship to biomedical, lifestyle, and cognitive factors. Neurobiol. Aging 2020, 92, 34–42.
- Sone, D.; Beheshti, I.; Shinagawa, S.; Niimura, H.; Kobayashi, N.; Kida, H.; Shikimoto, R.; Noda, Y.; Nakajima, S.; Bun, S.; et al. Neuroimaging-derived brain age is associated with life satisfaction in cognitively unimpaired elderly: A community-based study. Transl. Psychiatry 2022, 12, 25.
- Beheshti, I.; Maikusa, N.; Matsuda, H. The association between “Brain-Age Score”(BAS) and traditional neuropsychological screening tools in Alzheimer’s disease. Brain Behav. 2018, 8, e01020.
- Valizadeh, S.; Hänggi, J.; Mérillat, S.; Jäncke, L. Age prediction on the basis of brain anatomical measures. Hum. Brain Mapp. 2016, 38, 997–1008.
- Sone, D.; Beheshti, I.; Maikusa, N.; Ota, M.; Kimura, Y.; Sato, N.; Koepp, M.; Matsuda, H. Neuroimaging-based brain-age prediction in diverse forms of epilepsy: A signature of psychosis and beyond. Mol. Psychiatry 2019, 26, 825–834.
- Beheshti, I.; Mishra, S.; Sone, D.; Khanna, P.; Matsuda, H. T1-weighted MRI-driven brain age estimation in Alzheimer’s disease and Parkinson’s disease. Aging Dis. 2020, 11, 618.
- Cole, J.; Ritchie, S.J.; Bastin, M.; Hernández, M.C.V.; Maniega, S.M.; Royle, N.; Corley, J.; Pattie, A.; Harris, S.E.; Zhang, Q.; et al. Brain age predicts mortality. Mol. Psychiatry 2017, 23, 1385–1392.
- Beheshti, I.; Maikusa, N.; Matsuda, H. The accuracy of T1-weighted voxel-wise and region-wise metrics for brain age estimation. Comput. Methods Programs Biomed. 2022, 214, 106585.
- Goyal, M.S.; Blazey, T.M.; Su, Y.; Couture, L.E.; Durbin, T.J.; Bateman, R.J.; Benzinger, T.L.-S.; Morris, J.C.; Raichle, M.E.; Vlassenko, A.G. Persistent metabolic youth in the aging female brain. Proc. Natl. Acad. Sci. USA 2019, 116, 3251–3255.
- Beheshti, I.; Nugent, S.; Potvin, O.; Duchesne, S. Disappearing metabolic youthfulness in the cognitively impaired female brain. Neurobiol. Aging 2021, 101, 224–229.
- Nemmi, F.; Levardon, M.; Péran, P. Brain-age estimation accuracy is significantly increased using multishell free-water reconstruction. Hum. Brain Mapp. 2022, 43, 2365–2376.
- Beheshti, I.; Ganaie, M.A.; Paliwal, V.; Rastogi, A.; Razzak, I.; Tanveer, M. Predicting Brain Age Using Machine Learning Algorithms: A Comprehensive Evaluation. IEEE J. Biomed. Health Inform. 2021, 26, 1432–1440.
- Beheshti, I.; Nugent, S.; Potvin, O.; Duchesne, S. Bias-adjustment in neuroimaging-based brain age frameworks: A robust scheme. NeuroImage Clin. 2019, 24, 102063.
- Butler, E.R.; Chen, A.; Ramadan, R.; Le, T.T.; Ruparel, K.; Moore, T.M.; Satterthwaite, T.D.; Zhang, F.; Shou, H.; Gur, R.C.; et al. Pitfalls in brain age analyses. Hum. Brain Mapp. 2021, 42, 4092–4101.
- Le, T.T.; Kuplicki, R.T.; McKinney, B.A.; Yeh, H.-W.; Thompson, W.K.; Paulus, M.P.; Tulsa 1000 Investigators; Aupperle, R.L.; Bodurka, J.; Cha, Y.-H.; et al. A Nonlinear Simulation Framework Supports Adjusting for Age When Analyzing BrainAGE. Front. Aging Neurosci. 2018, 10, 317.
- Cole, J.H.; Poudel, R.P.; Tsagkrasoulis, D.; Caan, M.W.; Steves, C.; Spector, T.D.; Montana, G. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage 2017, 163, 115–124.
- Feng, X.; Lipton, Z.C.; Yang, J.; Small, S.A.; Provenzano, F.A.; Alzheimer’s Disease Neuroimaging Initiative; Australian Imaging Biomarkers and Lifestyle flagship study of ageing; Frontotemporal Lobar Degeneration Neuroimaging Initiative. Estimating brain age based on a uniform healthy population with deep learning and structural magnetic resonance imaging. Neurobiol. Aging 2020, 91, 15–25.
- Ning, K.; Duffy, B.A.; Franklin, M.; Matloff, W.; Zhao, L.; Arzouni, N.; Sun, F.; Toga, A.W. Improving brain age estimates with deep learning leads to identification of novel genetic factors associated with brain aging. Neurobiol. Aging 2021, 105, 199–204.
- Popescu, S.G.; Glocker, B.; Sharp, D.J.; Cole, J.H. Local Brain-Age: A U-Net Model. Front. Aging Neurosci. 2021, 13, 761954.
- Levakov, G.; Rosenthal, G.; Shelef, I.; Raviv, T.R.; Avidan, G. From a deep learning model back to the brain—Identifying regional predictors and their relation to aging. Hum. Brain Mapp. 2020, 41, 3235–3252.
