The Scribble polarity module is composed by Scribble (Scrib), Discs large 1 (Dlg1) and Lethal (2) giant larvae (L(2)gl), a group of highly conserved neoplastic tumor suppressor genes (TSGs) from flies to humans. Even though the Scribble module has been profusely studied in epithelial cell polarity, the number of tissues and processes in which it is involved is increasingly growing. Here we discuss the role of the Scribble module in the asymmetric division of Drosophila neuroblasts (NBs), as well as the underlying mechanisms by which those TSGs act in this process. Finally, we also describe what we know about the consequences of mutating these genes in impairing the process of asymmetric NB division and promoting tumor-like overgrowth.
- Scribble polarity module
- asymmetric cell division
- neuroblasts
- tumorigenesis
- Drosophila
1. Asymmetric Division of Drosophila Neuroblasts
NBs, the neural stem cells of the Drosophila central nervous system (CNS), divide asymmetrically to give rise to another NB that keeps on dividing and a daughter cell called ganglion mother cell (GMC) that will start a differentiation program [1][2][3]. This cell fate commitment is possible by the action of cell-fate determinants, which are asymmetrically located at the basal pole of metaphase NBs and segregate exclusively to the GMC during NB division (Figure 1). The translational regulator brain tumor (Brat), the transcription factor Prospero (Pros), and the cytoplasmic protein Numb are among those determinants that inhibit proliferation and activate differentiation in the GMC [4][5][6][7][8][9][10][11][12].
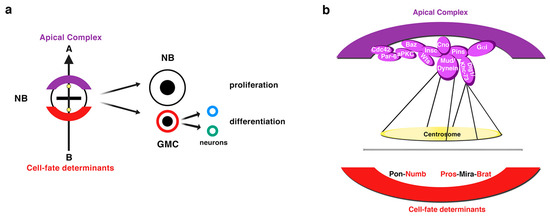
Figure 1. Drosophila neuroblasts (NBs), the neural stem cells of the central nervous system (CNS), divide asymmetrically. (a) NBs divide asymmetrically to give rise to another NB and a ganglion mother cell (GMC), which receives the cell-fate determinants that induce a differentiation program in this cell. The GMC divides asymmetrically through a terminal division to give rise to two different neurons of glial cells. The sibling NB that does not receive the cell-fate determinants keeps on dividing. A group of proteins apically located at the cortex of metaphase NBs (the “apical complex”) is in turn crucial for the basal sorting of the cell-fate determinants, as well as for the correct orientation of the mitotic spindle along an apico-basal axis of cell polarity previously established. (b) A diagram showing the most representative components of the apical complex and the cell-fate determinants Numb, Pros and Brat. Pon and Mira are adaptor proteins of Numb (Pon) and of Pros and Brat (Mira) (modified from Carmena, Fly, 2018).
A group of proteins located at the apical cortex of metaphase NBs control, in turn, the basal sorting of cell-fate determinants, as well as the orientation of the mitotic spindle along the NB apico-basal axis of polarity, two key processes to ensure the asymmetry of the division. This apical complex is an intricate protein network that includes the conserved partitioning defective proteins Par-6 and Par-3 (Bazooka, Baz, in Drosophila) and the atypical protein kinase C (aPKC) (Figure 1) [13][14][15][16][17]. Baz physically interacts with the adaptor protein Inscuteable (Insc) that in turn binds and activates Partner of Insc (Pins; LGN in mammals), allowing the interaction between the Gαi protein subunit anchored to the membrane and Pins, which thereafter orchestrates the orientation of the spindle (Figure 1) [18][19][20][21][22][23][24][25]. This process requires the function of Canoe (Cno; Afadin in mammals) that, after being phosphorylated by the serine-threonine kinase Warts (Wts; LATS1-2 in mammals), binds the N-terminal PinsTPR domain, the same region that Insc was bound to [25][26][27][28]. Cno then contributes to the apical recruitment of the Pins-interacting proteins Mushroom body defect (Mud; NuMA in mammals) and Dlg1 [26][27]. Dlg1 binds the middle PinsLINKER domain and the Kinesin heavy chain 73 (Khc-73) motor protein that interacts with astral microtubule plus-ends, anchoring the spindle to the apical cortex [29][30][31]. Mud, like Cno, interacts with the PinsTPR domain and, additionally, with the Dynein molecular motor, which binds the astral microtubule minus-ends promoting pulling forces on them and reinforcing the apical-basal orientation of the spindle [29] (Figure 1).
2. Types of Neuroblasts: Different Lineages, Same Origin
Embryonic NBs delaminate from the neuroectoderm inheriting the apico-basal polarity of the neuroepithelial cells. The establishment of an axis of cell polarity is a prerequisite for a correct asymmetric division. Once this axis of cell polarity is established, the mitotic spindle aligns along it and the cell-fate determinants localize asymmetrically at the basal pole of the NB. These embryonic NBs will divide a finite number of times, up to twenty, entering quiescence at the end of embryogenesis. At late first larval stage, NBs resume proliferation, this time undergoing hundreds of them and increasing their size before each division. These NBs that divide to give rise to another NB and a GMC have been called type I NBs (Figure 2) [2]. Some years ago, another type of NBs, called type II NBs, were found in the larval central brain [6][32][33]. These NBs also divide asymmetrically to give rise to another NB and, instead of a GMC, a progenitor cell called an intermediate progenitor (INP) that, after a maturation process, will divide asymmetrically to give rise to another INP and a GMC (Figure 2). Given this additional phase of proliferation, type II NB lineages are larger than type I and more prone to induce tumor-like overgrowth when the process of ACD is compromised (see below). In addition, while type I NB lineages occupy most of the central brain, these type II NB lineages are only eight per brain hemisphere and are located at precise locations at the dorso-medial part of the brain (Figure 2). Very recently, it has been shown that type II NBs have also an embryonic origin and are arrested at the end of embryogenesis [34][35].
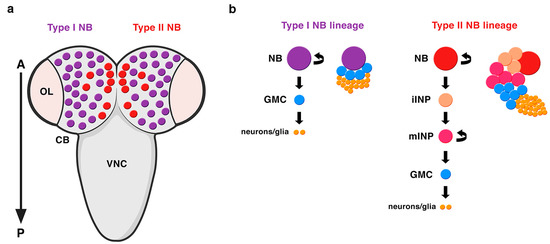
Figure 2. Types of NBs in the Drosophila CNS. (a) A dorsal view of the larval central brain (CB) containing type I (purple) and type II (red) NBs. There are only eight type II NB lineages per brain hemisphere located at very specific positions at the dorso-medial part of the CB. OL: optic lobe; VNC: ventral nerve cord; A: anterior; P: posterior. (b) Type II NB lineages are bigger than type I NB lineages. In type II NB lineages, the NB divides asymmetrically to generate another NB and, instead of a GMC (like in type I NB lineages), an intermediate progenitor (INP), which after a process of maturation, divides asymmetrically to give rise to another INP and a GMC. iINP: immature INP; mINP: mature INP (modified from Carmena, Fly, 2018).
The Scribble Module in Asymmetric Neuroblast Division during Development
A role for the neoplastic TSGs of the Scribble module in asymmetric NB division was first shown for Dlg1 and L(2)gl [36][37]. Dlg1 and L(2)gl were found to be essential for the basal targeting of the cell-fate determinants Numb and Pros, as well as of their adaptor proteins Partner of Numb (Pon) and Miranda (Mira), respectively, in both embryonic and larval mitotic NBs [36][37]. However, Dlg1 and L(2)gl were dispensable for the localization of apical proteins, such as Baz, Insc or Pins and for the orientation of the mitotic spindle [36][37]. Dlg1 was required for the cortical localization of L(2)gl, which became cytoplasmic in dlg1 mutant embryos; however, L(2)gl was not necessary for the localization of Dlg1. Hence, it was proposed that, at least for its localization, although not necessarily for its function, Dlg1 would act upstream of L(2)gl [36][37]. In fact, we now know that L(2)gl acts functionally upstream of Dlg1 [38]. Both proteins are distributed predominantly at the cortex, although, at metaphase, Dlg1 is apically enriched while L(2)gl is phosphorylated and inactivated by aPKC at this location [39]. This is promoted by Aurora-A (AurA) kinase, which at metaphase phosphorylates Par-6 with the consequent activation of aPKC. Activated aPKC phosphorylates and inactivates L(2)gl, which leaves the apical complex and it is replaced by Baz/Par-3 [38][39][40]. Baz, then, allows the phosphorylation of the cell-fate determinant Numb by aPKC, and the consequent exclusion of P-Numb to the basal pole of the NB [38] (Figure 3a). The inhibition of L(2)gl by aPKC is mutual, as L(2)gl represses aPKC basally, restricting it to the apical cortex [41] (Figure 3b, c). Thus, the localization of at least some apical proteins, such as aPKC, do depend on some of the TSGs of the Scribble module. L(2)gl also binds and represses non-muscle myosin II heavy chain, called Zipper in Drosophila, at interphase. At metaphase, when L(2)gl is inactivated by aPKC, it was proposed that myosin II becomes active and, in turn, promotes the cortical exclusion of the cell-fate determinant adaptor protein Mira from the apical NB cortex (Figure 3b) [42][43]. The basal targeting of Mira would occur by passive diffusion throughout the cytoplasm, not by active transport, and it would depend on another myosin, myosin VI, Jaguar in Drosophila, which would be essential for the final localization of Mira in a basal crescent (Figure 3b) [44][45]. Yet, the role of myosin II in Mira localization (Figure 3b) was questioned and the model to explain Mira asymmetry was replaced by another one some years ago [46]. This latter work showed that aPKC can directly phosphorylate Mira at several sites to exclude it from the apical cortex independently of L(2)gl, which would be antagonizing aPKC activity (Figure 3c) [40][46]. More recently, additional data seem to point to an integrated view of both models [47]. Thus, aPKC direct phosphorylation of Mira, event that occurs at prophase, would not be the only mechanism that regulates Mira asymmetry, and an actomyosin-dependent mechanism would be additionally required to maintain Mira asymmetric localization at metaphase (Figure 3d) [47][48].
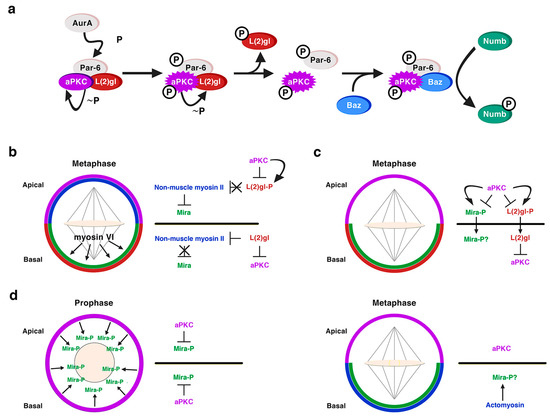
Figure 3. L(2)gl in asymmetric NB division. (a) L(2)gl forms part of an inactive Par complex. At metaphase, the kinase AurA phosphorylates Par6, which leads to the activation of aPKC and the consequent phosphorylation of L(2)gl by active aPKC. P-L(2)gl then leaves the Par complex and it is replaced by Baz/Par-3, which binds both aPKC and Numb making possible the phosphorylation of Numb by aPKC and the exclusion of P-Numb from the apical cortex. (Modified from Wirtz-Peitz et al., Cell, 2008). (b) Myosin-dependent model to explain the basal sorting of the adaptor protein Mira. aPKC phosphorylates and inactivates L(2)gl at the apical pole of metaphase NBs. Hence L(2)gl cannot bind and inactivate myosin II, which excludes Mira from the apical cortex. Myosin VI would help to locate Mira in a basal crescent. L(2)gl is active at the basal pole inhibiting both aPKC and myosin II, allowing in this way the accumulation of Mira at this location. (c) Myosin-independent model to explain the basal sorting of Mira. Apical aPKC directly phosphorylates both L(2)gl and Mira excluding them from the apical cortex. At the basal pole L(2)gl counteracts the activity of aPKC. (d) An integrative model both aPKC and myosin-dependent. At prophase, before the nuclear membrane is disorganized, cortical aPKC phosphorylates Mira and excludes it from the cortex. At metaphase, aPKC is apically enriched and the basal actomyosin network contributes to the asymmetric Mira retention by providing an anchoring scaffold to Mira at this location. The role of L(2)gl is not discussed in the context of this model (Hannaford et al. eLife, 2018), but it could be also counteracting the activity of aPKC basally.
Regarding Dlg1, over the past 20 years, since it was first described in the process of NB asymmetric division [36][37], we have substantially increased our knowledge about the mechanisms underlying Dlg1 function in this context. The guanylate kinase (GK) domain of Dlg1/DLG1 (Figure 4a), a phosphoprotein recognition motif, binds the Pins/LGN linker domain (PinsLinker) both in Drosophila and in mammals [29][49][50]. This conserved PinsLinker domain must be phosphorylated by the mitotic kinase AurA to physically interact with the Dlg1 GK domain [29], which in turn recruits the motor protein Khc-73. This kinase first interacts through its MAGUK binding stalk (MBS) domain (Khc-73MBS) with the GK motif of Dlg1 at the cortex, and then with astral microtubule plus-ends through its motor domain (Khc-73motor) (Figure 4a). This Pins-Dlg1-Khc73 pathway mediates a microtubule-induced Pins-Gαi (the latter is bound to the GoLoco domains of Pins, see above and Figure 1) cortical polarity at metaphase NBs, independently of the Par complex [30]. However, this pathway is not enough for a full orientation of the mitotic spindle. Pins must activate another microtubule motor pathway mediated by Dynein that interacts with minus-end astral microtubules. The PinsTPR domain is the motif involved in the activation of this pathway by binding Mud/NuMA, which in turn associates with Dynein that exerts pulling forces on microtubules. Both PinsTPR- and PinsLinker-mediated pathways are required and act synergistically to promote a robust spindle alignment [29]. The mechanism by which these Pins-mediated pathways interact was identified some years ago [51] (Figure 4b). In this work, authors show how the Drosophila 14-3-3ζ protein associates to the 14-3-3 binding motif present in the Khc-73 C-terminal stalk, (Figure 4a). The NudE Dynein cofactor [52][53] interacts in turn with 14-3-3ε, which forms a heterodimer with 14-3-3ζ. This complex 14-3-3ζ/14-3-3ε/NudE acts then as the bridge between both Pins-mediated pathways to allow a full, optimal spindle orientation (Figure 4b) [51]. More recently, Dlg1 has been shown to be phosphorylated in its SH3 domain by aPKC [54] (Figure 4c). This phosphorylation releases an auto-inhibitory intramolecular interaction between Dlg1 SH3 and the GK domains. In this situation, the spindle orientation factor Gukh can bind to the Dlg1 GK domain and to astral microtubules, contributing, along other Dlg1 effectors such as Khc-73, to Dlg1-mediated spindle alignment (Figure 4c).
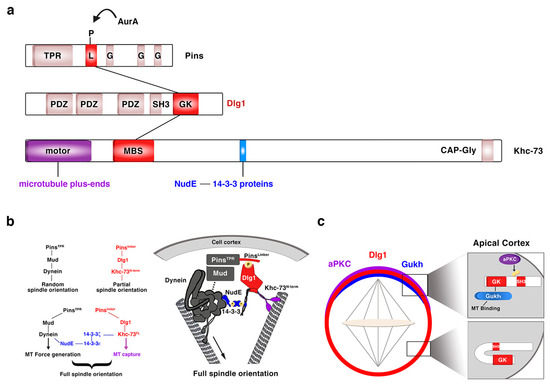
Figure 4. Dlg1 in asymmetric NB division. (a) Modular structure of the ACD regulators Pins, Dlg1 and Khc-73. The kinase AurA phosphorylates the linker domain (L) of Pins, and the GK domain of Dlg1 binds both this phosphorylated PinsLinker domain and the MBS motif of Khc-73. This kinase binds astral microtubule plus-ends through its motor domain and 14-3-3ζ protein through a 14-3-3 binding motif present at the C-terminal stalk, between the MBS and the CAP-Gly motif. TPR: TetratricoPeptide Repeat; L: Linker; G: GoLoco; PDZ: PSD-95/Dlg/ZO-1; SH3: Src Homology 3; GK: Guanylate Kinase; MBS: Maguk Binding Stalk; CAP-Gly: Cytoskeleton Associated Proteins-Glycine-rich. (b) The two Pins-mediated pathways that orientate the mitotic spindle are connected through a NudE-14-3-3 protein bridge, which binds the two motor proteins involved in each of those pathways. NudE binds the motor Dynein and 14-3-3ε, which forms a heterodimer with 14-3-3ζ that in turn interacts with the motor Khc-73 (adapted from Lu and Prehoda, Dev Cell, 2013). (c) aPKC phosphorylates the SH3 domain of Dlg1 releasing an intramolecular inhibitory binding between SH3 and GK domains. GK can then bind the microtubule interactor protein Gukh, which contributes to the proper orientation of the mitotic spindle (adapted from Golub et al., eLIFE, 2017).
As mentioned above, Scrib was identified later than L(2)gl and Dlg1 [55] and consequently, it was described to be involved in NB asymmetric division a posteriori than those ACD regulators . In this work, Scrib localization was found to be cortical in NBs, with an apical enrichment at metaphase, similar to Dlg1 distribution. Likewise, as L(2)gl, Scrib localization was dependent on Dlg1 [56]. Authors described for the first time the function of all these TSGs, L(2)gl, Dlg1 and Scrib, in regulating cell size and mitotic spindle asymmetry in NBs. While in wild-type telophase NBs, the NB was bigger than the GMC, and the apical centrosome and astral microtubules larger than the basal ones, in l(2)gl, dlg1 and scrib embryonic mutant NBs, symmetric divisions (with equal-sized NB and GMC) and even inverted divisions (with the NB smaller than the GMC) were detected [56]. Scrib, as previously shown for L(2)gl and Dlg1, was found to be required for basal targeting of cell fate determinants and adaptor proteins, such as Mira and Pros, but not for the localization of apical proteins [56]. More recently, however, the apical protein aPKC has been shown to require Scrib for a proper cortical crescent formation at metaphase in type II NB lineages of the larval brain [57]. Thus, over the past years, all of these TSGs (L(2)gl, Dlg1, and Scrib) have been shown to be also necessary for the correct localization of at least some apical proteins (i.e., L(2)gl for aPKC; Dlg1 for Pins and Scrib for aPKC localization). Some of the Scrib motifs, such as the LRR region and the PDZ domains, have been proved to be required for the proper cortical localization and function of Scrib in NBs [58]. However, while the mechanisms by which L(2)gl and Dlg1 regulate the asymmetric division of NBs have been deeply investigated over the past years, we do not have any clear clue about the underlying mechanisms or mode of action of Scrib in this context.
3. The Scribble Module in Asymmetric Neuroblast Division during Tumorigenesis
ACD is a fundamental process during development to generate cell diversity. In addition, as we have learned over the past years, ACD is also a relevant process to take into account in the context of cancer and stem cell biology. A connection between failures in the process of ACD and tumorigenesis was first shown in the lab of C. González using the neural stem cells or NBs of the Drosophila larval brain as a model system [59]. In these experiments, pieces of GFP-labeled brains mutant for different ACD regulators were transplanted into the abdomen of adult host flies. These flies, after several weeks, developed big tumoral masses inside their abdomen, tumors that in some cases induced metastatic growth [59]. However, mutations in genes involved in ACD modulation do not always cause tumor-like overgrowth. It will depend on the type of ACD regulator and the particular environment in which the NB lineage grows [60]. For example, type II NB clones in the larval brain mutant for the ACD regulator gene cno/AFDN or for each of the Scribble module (l(2)gl, dlg1 and scrib) do show ectopic NBs within the clone but they do not overgrow [57]. In fact, at least the scrib mutant clones are smaller than control NB clones and they do not appear very frequently. As it occurs in epithelial scrib mutant clones, in scrib NB clones a JNK activity-dependent apoptosis is also triggered [57]. However, the simultaneous loss of scrib and cno/AFDN in these larval NB clones overcomes the scrib/JNK-induced apoptosis and causes massive tumor-like overgrowths [57]. This effect is due to the upregulation of Ras, normally repressed by Cno/Afadin [61][62]. Activated Ras, then, promotes a switch in the JNK function, from a pro-apoptotic to a pro-growth effect, similar to what occurs in epithelial RasV12 scrib double mutant clones [63][64][65][57]. Neither cno l(2)gl nor cno dlg1 double mutant clones show the strong synergistic cooperation displayed in cno scrib mutant clones. In fact, the former double mutant clones are very similar to cno single mutant clones [57]. One possibility to explain the different behavior of cno l(2)gl and cno dlg1 mutant clones is that JNK is not activated in l(2)gl nor in dlg1 NB single mutant clones, even though in epithelia JNK is activated in each of those single mutant clones [64]. This is something that should be analyzed in detail in NB mutant clones. Nevertheless, the capability or not of inducing JNK in the l(2)gl or dlg1 single NB mutant clones is probably not the only explanation, as RasV12 scrib NB mutant clones do not show the tumor-like overgrowth shown by cno scrib NB mutant clones [57]. Thus, altogether, the data we currently have strongly suggest that Cno is acting in the same pathway that the ACD regulators Dlg1 and L(2)gl and is epistatic to them. This is consistent with previous results showing that Cno contributes to Dlg1 recruitment to the apical pole of the NB [26] and that Cno is required for a proper aPKC cortical localization [57], as aPKC acts upstream of L(2)gl [38]. However, Scrib must be working in at least a partially independent pathway to that involving the ACD regulators, Cno, L(2)gl, and Dlg1, and this would explain the strongest effect of cno scrib double mutant clones. Hence, in asymmetric NB division, the Scribble module does not seem to be so functionally interdependent as in epithelia.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21082865
References
- Chris Q. Doe; Neural stem cells: balancing self-renewal with differentiation. Development 2008, 135, 1575-1587, 10.1242/dev.014977.
- Emmanuel Gallaud; Tri Pham; Clemens Cabernard; Drosophila melanogaster Neuroblasts: A Model for Asymmetric Stem Cell Divisions.. Results and Problems in Cell Differentiation 2017, 61, 183-210, 10.1007/978-3-319-53150-2_8.
- Zsolt G. Venkei; Yukiko M Yamashita; Emerging mechanisms of asymmetric stem cell division. Journal of Cell Biology 2018, 217, 3785-3795, 10.1083/jcb.201807037.
- Bruno Bello; Heinrich Reichert; Frank Hirth; The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 2006, 133, 2639-2648, 10.1242/dev.02429.
- Joerg Betschinger; Karl Mechtler; Juergen A. Knoblich; Asymmetric Segregation of the Tumor Suppressor Brat Regulates Self-Renewal in Drosophila Neural Stem Cells. Cell 2006, 124, 1241-1253, 10.1016/j.cell.2006.01.038.
- Sarah K. Bowman; Vivien Rolland; Joerg Betschinger; Kaolin A. Kinsey; Gregory Emery; Juergen A. Knoblich; The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila. Developmental Cell 2008, 14, 535-46, 10.1016/j.devcel.2008.03.004.
- Asymmetric segregation of the homeodomain protein PROSPERO during Drosophila development. Trends in Genetics 1996, 12, 46, 10.1016/0168-9525(96)90053-0.
- Asymmetric segregation of NUMB and PROSPERO during cell division. Trends in Genetics 1996, 12, 46, 10.1016/0168-9525(96)81395-3.
- Cheng-Yu Lee; Brian D. Wilkinson; Sarah E. Siegrist; Robin P. Wharton; Chris Q. Doe; Brat Is a Miranda Cargo Protein that Promotes Neuronal Differentiation and Inhibits Neuroblast Self-Renewal. Developmental Cell 2006, 10, 441-449, 10.1016/j.devcel.2006.01.017.
- Michelle S. Rhyu; Lily Yeh Jan; Yuh Nung Jan; Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 1994, 76, 477-491, 10.1016/0092-8674(94)90112-0.
- The PROSPERO transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Trends in Genetics 1996, 12, 46, 10.1016/0168-9525(96)81394-1.
- Tadashi Uemura; Susan Shepherd; Larry Ackerman; Lily Yeh Jan; Yuh Nung Jan; numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 1989, 58, 349-360, 10.1016/0092-8674(89)90849-0.
- Ute Kuchinke; Ferdi Grawe; Elisabeth Knust; Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka.. Current Biology 1999, 8, 1357-1365, 10.1016/s0960-9822(98)00016-5.
- Mark Petronczki; Juergen A. Knoblich; DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nature 2000, 3, 43-49, 10.1038/35050550.
- Markus Schober; Matthias R. Schaefer; Juergen A. Knoblich; Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 1999, 402, 548-551, 10.1038/990135.
- Andreas Wodarz; Andreas Ramrath; Alexandra Grimm; Elisabeth Knust; Drosophila Atypical Protein Kinase C Associates with Bazooka and Controls Polarity of Epithelia and Neuroblasts. Journal of Cell Biology 2000, 150, 1361-1374, 10.1083/jcb.150.6.1361.
- Andreas Wodarz; Andreas Ramrath; Ute Kuchinke; Elisabeth Knust; Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 1999, 402, 544-547, 10.1038/990128.
- Simone Culurgioni; Andrea Alfieri; Valentina Pendolino; Federica Laddomada; Marina Mapelli; Inscuteable and NuMA proteins bind competitively to Leu-Gly-Asn repeat-enriched protein (LGN) during asymmetric cell divisions. Proceedings of the National Academy of Sciences 2011, 108, 20998-21003, 10.1073/pnas.1113077108.
- Rachel Kraut; Jose A. Campos-Ortega; inscuteable,A Neural Precursor Gene ofDrosophila,Encodes a Candidate for a Cytoskeleton Adaptor Protein. Developmental Biology 1996, 174, 65-81, 10.1006/dbio.1996.0052.
- Rachel Kraut; William Chia; Lily Yeh Jan; Yuh Nung Jan; Juergen A. Knoblich; Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 1996, 383, 50-55, 10.1038/383050a0.
- Marie-Laure Parmentier; D Woods; S Greig; P G Phan; A Radovic; P Bryant; C J O'kane; Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila.. The Journal of Neuroscience 2000, 20, , .
- Matthias R. Schaefer; Mark Petronczki; Daniela Dorner; Michael Forte; Juergen A. Knoblich; Heterotrimeric G Proteins Direct Two Modes of Asymmetric Cell Division in the Drosophila Nervous System. Cell 2001, 107, 183-194, 10.1016/s0092-8674(01)00521-9.
- M Schaefer; A Shevchenko; J A Knoblich; A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila.. Current Biology 2000, 10, , .
- Yu, F.; Cai, Y.; Kaushik, R.; Yang, X.; Chia, W. Distinct roles of Galphai and Gbeta13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J. Cell Biol. 2003, 162, 623–633, doi:10.1083/jcb.200303174.
- Fengwei Yu; Xavier Morin; Yu Cai; Xiaohang Yang; William Chia; Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization.. Cell 2000, 100, 399-409, 10.1016/s0092-8674(00)80676-5.
- Alyona Keder; Noemí Rives-Quinto; Birgit L. Aerne; Maribel Franco; Nicolas Tapon; Ana Carmena; The Hippo Pathway Core Cassette Regulates Asymmetric Cell Division. Current Biology 2015, 25, 2739-2750, 10.1016/j.cub.2015.08.064.
- Stephan Speicher; Anja Fischer; Juergen A. Knoblich; Ana Carmena; The PDZ Protein Canoe Regulates the Asymmetric Division of Drosophila Neuroblasts and Muscle Progenitors. Current Biology 2008, 18, 831-837, 10.1016/j.cub.2008.04.072.
- Brett Wee; Christopher A. Johnston; Kenneth E. Prehoda; Chris Q. Doe; Canoe binds RanGTP to promote PinsTPR/Mud-mediated spindle orientation. Journal of Cell Biology 2011, 195, 369-376, 10.1083/jcb.201102130.
- Christopher A. Johnston; Keiko Hirono; Kenneth E. Prehoda; Chris Q. Doe; Identification of an Aurora-A/PinsLINKER/ Dlg Spindle Orientation Pathway using Induced Cell Polarity in S2 Cells. Cell 2009, 138, 1150-63, 10.1016/j.cell.2009.07.041.
- Sarah E. Siegrist; Chris Q. Doe; Microtubule-Induced Pins/Gαi Cortical Polarity in Drosophila Neuroblasts. Cell 2005, 123, 1323-1335, 10.1016/j.cell.2005.09.043.
- Kaori H. Yamada; Toshihiko Hanada; Athar H. Chishti; The Effector Domain of Human Dlg Tumor Suppressor Acts as a Switch That Relieves Autoinhibition of Kinesin-3 Motor GAKIN/KIF13B†. Biochemistry 2007, 46, 10039-10045, 10.1021/bi701169w.
- Jason Q. Boone; Chris Q. Doe; Identification ofDrosophilatype II neuroblast lineages containing transit amplifying ganglion mother cells. Developmental Neurobiology 2008, 68, 1185-95, 10.1002/dneu.20648.
- Bruno Bello; Natalya Izergina; Emmanuel Caussinus; Heinrich Reichert; Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Development 2008, 3, 5-5, 10.1186/1749-8104-3-5.
- José-Andrés Álvarez; Fernando Diaz-Benjumea; Origin and specification of type II neuroblasts in theDrosophilaembryo. Development 2018, 145, dev158394, 10.1242/dev.158394.
- Kathleen T. Walsh; Chris Q. Doe; Drosophila embryonic type II neuroblasts: origin, temporal patterning, and contribution to the adult central complex. 2017, , 170043, 10.1101/170043.
- Tomokazu Ohshiro; Takako Yagami; Chuan Zhang; Fumio Matsuzaki; Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 2000, 408, 593-596, 10.1038/35046087.
- Chian-Yu Peng; Laurina Manning; Roger Albertson; Chris Q. Doe; The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 2000, 408, 596-600, 10.1038/35046094.
- Frederik Wirtz-Peitz; Takashi Nishimura; Juergen A. Knoblich; Linking Cell Cycle to Asymmetric Division: Aurora-A Phosphorylates the Par Complex to Regulate Numb Localization. Cell 2008, 135, 161-73, 10.1016/j.cell.2008.07.049.
- Jörg Betschinger; Karl Mechtler; Juergen A. Knoblich; The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 2003, 422, 326-330, 10.1038/nature01486.
- Matthew J. Bailey; Kenneth E. Prehoda; Establishment of Par-Polarized Cortical Domains via Phosphoregulated Membrane Motifs.. Developmental Cell 2015, 35, 199-210, 10.1016/j.devcel.2015.09.016.
- Cheng-Yu Lee; Kristin J. Robinson; Chris Q. Doe; Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 2005, 439, 594-598, 10.1038/nature04299.
- D Strand; R Jakobs; G Merdes; B Neumann; A Kalmes; H W Heid; I Husmann; B M Mechler; The Drosophila lethal(2)giant larvae tumor suppressor protein forms homo-oligomers and is associated with nonmuscle myosin II heavy chain.. Journal of Cell Biology 1994, 127, 1361-1373, 10.1083/jcb.127.5.1361.
- Claudia Barros; Chris B. Phelps; Andrea H. Brand; Drosophila Nonmuscle Myosin II Promotes the Asymmetric Segregation of Cell Fate Determinants by Cortical Exclusion Rather Than Active Transport. Developmental Cell 2003, 5, 829-840, 10.1016/s1534-5807(03)00359-9.
- Veronika Erben; Markus Waldhuber; Diana Langer; Ingrid Fetka; Ralf Peter Jansen; Claudia Petritsch; Asymmetric localization of the adaptor protein Miranda in neuroblasts is achieved by diffusion and sequential interaction of Myosin II and VI. Journal of Cell Science 2008, 121, 1403-1414, 10.1242/jcs.020024.
- Claudia Petritsch; Gaia Tavosanis; Christoph W. Turck; Lily Yeh Jan; Yuh Nung Jan; The Drosophila Myosin VI Jaguar Is Required for Basal Protein Targeting and Correct Spindle Orientation in Mitotic Neuroblasts. Developmental Cell 2003, 4, 273-281, 10.1016/s1534-5807(03)00020-0.
- Scott X. Atwood; Kenneth E. Prehoda; aPKC Phosphorylates Miranda to Polarize Fate Determinants during Neuroblast Asymmetric Cell Division. Current Biology 2009, 19, 723-9, 10.1016/j.cub.2009.03.056.
- Matthew Hannaford; Anne Ramat; Nicolas Loyer; Jens Januschke; aPKC-mediated displacement and actomyosin-mediated retention polarize Miranda in Drosophila neuroblasts. eLife 2018, 7, , 10.7554/eLife.29939.
- Matthew Hannaford; Nicolas Loyer; Francesca Tonelli; Martin Zoltner; Jens Januschke; A chemical-genetics approach to study the role of atypical Protein Kinase C in Drosophila.. Development 2019, 146, dev170589, 10.1242/dev.170589.
- Jinwei Zhu; Yuan Shang; Caihao Xia; Wenning Wang; Wenyu Wen; Mingjie Zhang; Guanylate kinase domains of the MAGUK family scaffold proteins as specific phospho-protein-binding modules. The EMBO Journal 2011, 30, 4986-4997, 10.1038/emboj.2011.428.
- Christopher A. Johnston; Chris Q. Doe; Kenneth E. Prehoda; Structure of an Enzyme-Derived Phosphoprotein Recognition Domain. PLOS ONE 2012, 7, e36014, 10.1371/journal.pone.0036014.
- Michelle S. Lu; Kenneth E. Prehoda; A NudE/14-3-3 pathway coordinates dynein and the kinesin Khc73 to position the mitotic spindle.. Developmental Cell 2013, 26, 369-80, 10.1016/j.devcel.2013.07.021.
- Catherine Johnson; Sandra Crowther; Margaret J. Stafford; David G. Campbell; Rachel Toth; Carol Mackintosh; Bioinformatic and experimental survey of 14-3-3-binding sites. Biochemical Journal 2010, 427, 69-78, 10.1042/bj20091834.
- Vaishnavi Ananthanarayanan; Activation of the motor protein upon attachment: Anchors weigh in on cytoplasmic dynein regulation. BioEssays 2016, 38, 514-525, 10.1002/bies.201600002.
- Ognjen Golub; Brett Wee; Rhonda A Newman; Nicole M Paterson; Kenneth E. Prehoda; Author response: Activation of Discs large by aPKC aligns the mitotic spindle to the polarity axis during asymmetric cell division. Author response 2017, 6, , 10.7554/elife.32137.018.
- D. Bilder; Cooperative Regulation of Cell Polarity and Growth by Drosophila Tumor Suppressors. Science 2000, 289, 113-116, 10.1126/science.289.5476.113.
- Roger Albertson; Chris Q. Doe; Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nature 2003, 5, 166-170, 10.1038/ncb922.
- Rives-Quinto, N.; Franco, M.; de Torres-Jurado, A.; Carmena, A. Synergism between canoe and scribble mutations causes tumor-like overgrowth via Ras activation in neural stem cells and epithelia. Development 2017, 144, 2570–2583, doi:10.1242/dev.148171.
- R. Albertson; Chiswili Chabu; Amy Sheehan; Chris Q. Doe; Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. Journal of Cell Science 2004, 117, 6061-6070, 10.1242/jcs.01525.
- Emmanuel Caussinus; Cayetano Gonzalez; Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nature Genetics 2005, 37, 1125-1129, 10.1038/ng1632.
- Ana Carmena; Compromising asymmetric stem cell division in Drosophila central brain: Revisiting the connections with tumorigenesis. Fly 2018, 12, 71-80, 10.1080/19336934.2017.1416277.
- Ana Carmena; Stephan Speicher; Mary Baylies; The PDZ Protein Canoe/AF-6 Links Ras-MAPK, Notch and Wingless/Wnt Signaling Pathways by Directly Interacting with Ras, Notch and Dishevelled. PLOS ONE 2006, 1, e66, 10.1371/journal.pone.0000066.
- G Radziwill; R A Erdmann; U Margelisch; K Moelling; The Bcr kinase downregulates Ras signaling by phosphorylating AF-6 and binding to its PDZ domain.. Molecular and Cellular Biology 2003, 23, , .
- Anthony M. Brumby; Helena E. Richardson; scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. The EMBO Journal 2003, 22, 5769-5779, 10.1093/emboj/cdg548.
- Tatsushi Igaki; Raymond A. Pagliarini; Tian Xu; Loss of Cell Polarity Drives Tumor Growth and Invasion through JNK Activation in Drosophila. Current Biology 2006, 16, 1139-1146, 10.1016/j.cub.2006.04.042.
- R. A. Pagliarini; Tian Xu; A Genetic Screen in Drosophila for Metastatic Behavior. Science 2003, 302, 1227-1231, 10.1126/science.1088474.
