SARS-CoV-2 mainly targets the respiratory tract, resulting in rapid and severe respiratory symptoms and lung failure, as well as some clinical symptoms such as fever, dry cough, fatigue, and dyspnea. In addition, studies reported gastrointestinal disturbances such as loss of appetite followed by diarrhea, nausea, and abdominal pain in the infected patients. Indeed, SARS-CoV-2 can also acutely replicate in the mucosa of the patient’s small intestine and excrete its RNA into the patient’s stool. In addition to these manifestations, many patients have exhibited a variety of symptoms (e.g., olfactory and gustatory disturbances, anosmia, headache, dysgeusia, confusion, and fatigue), which could be attributed to cranial nerve involvement. SARS-CoV-2, as with other coronaviruses, may initially invade peripheral-nerve endings and then progress regularly to the central nervous system via synaptic-connected junctions. Microalgae are eukaryotic, microscopic, and photosynthetic lower organisms that have recently been considered a more promising platform for the production of various biologics, especially complex glycosylated proteins.
- ACE2
- biopharmaceuticals
- edible vaccine
- COVID-19
- microalgae
- SARS-CoV-2
- S-glycoprotein
1. Therapeutic Potential of SARS-CoV-2 S-Glycoprotein
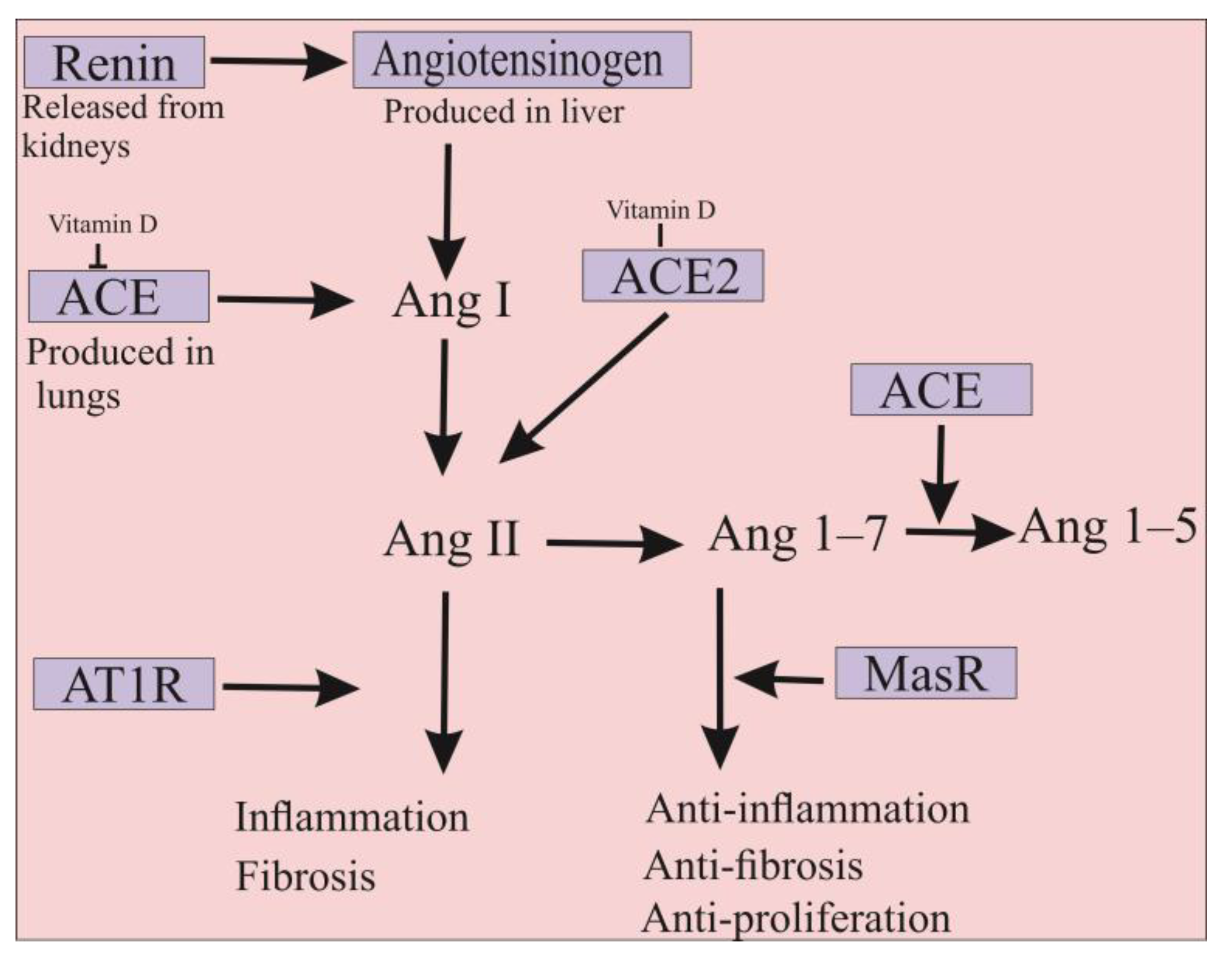
2. ACE2 Is Also a Promising Approach for the Treatment of COVID-19 Infection
3. Potential of Microalgae for the Production of Recombinant S-Glycoprotein and ACE2
This entry is adapted from the peer-reviewed paper 10.3390/md20110657
References
- Martínez-Flores, D.; Zepeda-Cervantes, J.; Cruz-Reséndiz, A.; Aguirre-Sampieri, S.; Sampieri, A.; Vaca, L. SARS-CoV-2 Vaccines Based on the Spike Glycoprotein and Implications of New Viral Variants. Front. Immunol. 2021, 12, 701501.
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons from the Past. Front. Immunol. 2020, 11, 1949.
- Singh, L.; Bajaj, S.; Gadewar, M.; Verma, N.; Ansari, M.N.; Saeedan, A.S.; Kaithwas, G.; Singh, M. Modulation of Host Immune Response Is an Alternative Strategy to Combat SARS-CoV-2 Pathogenesis. Front. Immunol. 2021, 12, 660632.
- Pourseif, M.M.; Parvizpour, S.; Jafari, B.; Dehghani, J.; Naghili, B.; Omidi, Y. Prophylactic domain-based vaccine against SARSCoV-2, causative agent of COVID-19 pandemic. BioImpacts 2020, 11, 65–84.
- Xia, X. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses 2021, 13, 109.
- Almehdi, A.M.; Khoder, G.; Alchakee, A.S.; Alsayyid, A.T.; Sarg, N.H.; Soliman, S.S.M. SARS-CoV-2 spike protein: Pathogenesis, vaccines, and potential therapies. Infection 2021, 49, 855–876.
- Gong, Y.; Qin, S.; Dai, L.; Tian, Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct. Target. Ther. 2021, 6, 396.
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236.
- Akash, K.; Sharma, A.; Kumar, D.; Singh, S.K.; Gupta, G.; Chellappan, D.K.; Dua, K.; Nagraik, R. Molecular aspects of Omicron, vaccine development, and recombinant strain XE: A review. J. Med. Virol. 2022, 94, 4628–4643.
- Wang, N.; Shang, J.; Jiang, S.; Du, L. Subunit Vaccines Against Emerging Pathogenic Human Coronaviruses. Front. Microbiol. 2020, 11, 298.
- Kam, Y.W.; Kien, F.; Roberts, A.; Cheung, Y.C.; Lamirande, E.W.; Vogel, L.; Chu, S.L.; Tse, J.; Guarner, J.; Zaki, S.R.; et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine 2007, 25, 729–740.
- Di Domenico, M.; De Rosa, A.; Boccellino, M. Detection of SARS-COV-2 Proteins Using an ELISA Test. Diagnostics 2021, 11, 698.
- Lee, C.Y.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11, 879.
- Mathuria, J.P.; Yadav, R.; Rajkumar. Laboratory diagnosis of SARS-CoV-2—A review of current methods. J. Infect. Public Health 2020, 13, 901–905.
- Xiao, L.; Sakagami, H.; Miwa, N. ACE2: The key Molecule for Understanding the Pathophysiology of Severe and Critical Conditions of COVID-19: Demon or Angel? Viruses 2020, 12, 491.
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D.; et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248.
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436, 112–116.
- Roca-Ho, H.; Riera, M.; Palau, V.; Pascual, J.; Soler, M.J. Characterization of ACE and ACE2 Expression within Different Organs of the NOD Mouse. Int. J. Mol. Sci. 2017, 18, 563.
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000, 275, 33238–33243.
- Sanchis-Gomar, F.; Lavie, C.J.; Perez-Quilis, C.; Henry, B.M.; Lippi, G. Angiotensin-Converting Enzyme 2 and Antihypertensives (Angiotensin Receptor Blockers and Angiotensin-Converting Enzyme Inhibitors) in Coronavirus Disease 2019. Mayo. Clin. Proc. 2020, 95, 1222–1230.
- Hamming, I.; Cooper, M.E.; Haagmans, B.L.; Hooper, N.M.; Korstanje, R.; Osterhaus, A.D.; Timens, W.; Turner, A.J.; Navis, G.; van Goor, H. The emerging role of ACE2 in physiology and disease. J. Pathol. 2007, 212, 1–11.
- Hamming, I.; van Goor, H.; Turner, A.J.; Rushworth, C.A.; Michaud, A.A.; Corvol, P.; Navis, G. Differential regulation of renal angiotensin-converting enzyme (ACE) and ACE2 during ACE inhibition and dietary sodium restriction in healthy rats. Exp. Physiol. 2008, 93, 631–638.
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637.
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care. Med. 2020, 46, 586–590.
- Kim, J.; Mukherjee, A.; Nelson, D.; Jozic, A.; Sahay, G. Rapid generation of circulating and mucosal decoy ACE2 using mRNA nanotherapeutics for the potential treatment of SARS-CoV-2. bioRxiv 2020.
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273.
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005, 11, 875–879.
- Yang, X.H.; Deng, W.; Tong, Z.; Liu, Y.X.; Zhang, L.F.; Zhu, H.; Gao, H.; Huang, L.; Liu, Y.L.; Ma, C.M.; et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007, 57, 450–459.
- Zhang, L.; Dutta, S.; Xiong, S.; Chan, M.; Chan, K.K.; Fan, T.M.; Bailey, K.L.; Lindeblad, M.; Cooper, L.M.; Rong, L.; et al. Engineered ACE2 decoy mitigates lung injury and death induced by SARS-CoV-2 variants. Nat. Chem. Biol. 2022, 18, 342–351.
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8.
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20.
- Pang, X.; Cui, Y.; Zhu, Y. Recombinant human ACE2: Potential therapeutics of SARS-CoV-2 infection and its complication. Acta. Pharmacol. Sin. 2020, 41, 1255–1257.
- Haschke, M.; Schuster, M.; Poglitsch, M.; Loibner, H.; Salzberg, M.; Bruggisser, M.; Penninger, J.; Krähenbühl, S. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin. Pharmacokinet. 2013, 52, 783–792.
- Khan, A.; Benthin, C.; Zeno, B.; Albertson, T.E.; Boyd, J.; Christie, J.D.; Hall, R.; Poirier, G.; Ronco, J.J.; Tidswell, M.; et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care 2017, 21, 234.
- Liu, P.; Wysocki, J.; Souma, T.; Ye, M.; Ramirez, V.; Zhou, B.; Wilsbacher, L.D.; Quaggin, S.E.; Batlle, D.; Jin, J. Novel ACE2-Fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney Int. 2018, 94, 114–125.
- Zhang, Z.; Zeng, E.; Zhang, L.; Wang, W.; Jin, Y.; Sun, J.; Huang, S.; Yin, W.; Dai, J.; Zhuang, Z.; et al. Potent prophylactic and therapeutic efficacy of recombinant human ACE2-Fc against SARS-CoV-2 infection in vivo. Cell Discov. 2021, 7, 65.
- Dehghani, J.; Adibkia, K.; Movafeghi, A.; Pourseif, M.M.; Omidi, Y. Designing a new generation of expression toolkits for engineering of green microalgae; robust production of human interleukin-2. Bioimpacts 2020, 10, 259–268.
- Hempel, F.; Lau, J.; Klingl, A.; Maier, U.G. Algae as protein factories: Expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS ONE 2011, 6, e28424.
- Hempel, F.; Maier, U.G. An engineered diatom acting like a plasma cell secreting human IgG antibodies with high efficiency. Microb. Cell Factories 2012, 11, 126.
- Vanier, G.; Hempel, F.; Chan, P.; Rodamer, M.; Vaudry, D.; Maier, U.G.; Lerouge, P.; Bardor, M. Biochemical Characterization of Human Anti-Hepatitis B Monoclonal Antibody Produced in the Microalgae Phaeodactylum tricornutum. PLoS ONE 2015, 10, e0139282.
- Vanier, G.; Stelter, S.; Vanier, J.; Hempel, F.; Maier, U.G.; Lerouge, P.; Ma, J.; Bardor, M. Alga-Made Anti-Hepatitis B Antibody Binds to Human Fcγ Receptors. Biotechnol. J. 2018, 13, e1700496.
- Mathieu-Rivet, E.; Mati-Baouche, N.; Walet-Balieu, M.L.; Lerouge, P.; Bardor, M. N- and O-Glycosylation Pathways in the Microalgae Polyphyletic Group. Front. Plant Sci. 2020, 11, 609993.
- Grama, S.B.; Liu, Z.; Li, J. Emerging Trends in Genetic Engineering of Microalgae for Commercial Applications. Mar. Drugs 2022, 20, 285.
- Slattery, S.S.; Giguere, D.J.; Stuckless, E.E.; Shrestha, A.; Briere, L.K.; Galbraith, A.; Reaume, S.; Boyko, X.; Say, H.H.; Browne, T.S.; et al. Phosphate-regulated expression of the SARS-CoV-2 receptor-binding domain in the diatom Phaeodactylum tricornutum for pandemic diagnostics. Sci. Rep. 2022, 12, 7010.
- Dehghani, J.; Adibkia, K.; Movafeghi, A.; Maleki-Kakelar, H.; Saeedi, N.; Omidi, Y. Towards a new avenue for producing therapeutic proteins: Microalgae as a tempting green biofactory. Biotechnol. Adv. 2020, 40, 107499.
- Hempel, F.; Maurer, M.; Brockmann, B.; Mayer, C.; Biedenkopf, N.; Kelterbaum, A.; Becker, S.; Maier, U.G. From hybridomas to a robust microalgal-based production platform: Molecular design of a diatom secreting monoclonal antibodies directed against the Marburg virus nucleoprotein. Microb. Cell Factories 2017, 16, 131.
- Eichler-Stahlberg, A.; Weisheit, W.; Ruecker, O.; Heitzer, M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 2009, 229, 873–883.
- Kim, D.H.; Kim, Y.T.; Cho, J.J.; Bae, J.H.; Hur, S.B.; Hwang, I.; Choi, T.J. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol. 2002, 4, 63–73.
- Ratha, S.K.; Renuka, N.; Rawat, I.; Bux, F. Prospective options of algae-derived nutraceuticals as supplements to combat COVID-19 and human coronavirus diseases. Nutrition 2021, 83, 111089.
- Wan Afifudeen, C.L.; Teh, K.Y.; Cha, T.S. Bioprospecting of microalgae metabolites against cytokine storm syndrome during COVID-19. Mol. Biol. Rep. 2022, 49, 1475–1490.
- Wannathong, T.; Waterhouse, J.C.; Young, R.E.; Economou, C.K.; Purton, S. New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2016, 100, 5467–5477.
- Lu, L.; Zhang, H.; Zhan, M.; Jiang, J.; Yin, H.; Dauphars, D.J.; Li, S.Y.; Li, Y.; He, Y.W. Preventing Mortality in COVID-19 Patients: Which Cytokine to Target in a Raging Storm? Front. Cell Dev. Biol. 2020, 8, 677.
- Ávila-Román, J.; Talero, E.; de Los Reyes, C.; García-Mauriño, S.; Motilva, V. Microalgae-derived oxylipins decrease inflammatory mediators by regulating the subcellular location of NFκB and PPAR-γ. Pharmacol. Res. 2018, 128, 220–230.
- Talukdar, J.; Bhadra, B.; Dattaroy, T.; Nagle, V.; Dasgupta, S. Potential of natural astaxanthin in alleviating the risk of cytokine storm in COVID-19. Biomed. Pharmacother. 2020, 132, 110886.
- Tzachor, A.; Rozen, O.; Khatib, S.; Jensen, S.; Avni, D. Photosynthetically Controlled Spirulina, but Not Solar Spirulina, Inhibits TNF-α Secretion: Potential Implications for COVID-19-Related Cytokine Storm Therapy. Mar. Biotechnol. 2021, 23, 149–155.
- Chia, W.Y.; Kok, H.; Chew, K.W.; Low, S.S.; Show, P.L. Can algae contribute to the war with Covid-19? Bioengineered 2021, 12, 1226–1237.
- Khavari, F.; Saidijam, M.; Taheri, M.; Nouri, F. Microalgae: Therapeutic potentials and applications. Mol. Biol. Rep. 2021, 48, 4757–4765.
- Guzmán, S.; Gato, A.; Lamela, M.; Freire-Garabal, M.; Calleja, J.M. Anti-inflammatory and immunomodulatory activities of polysaccharide from Chlorella stigmatophora and Phaeodactylum tricornutum. Phytother. Res. 2003, 17, 665–670.
- Kwon, P.S.; Oh, H.; Kwon, S.J.; Jin, W.; Zhang, F.; Fraser, K.; Hong, J.J.; Linhardt, R.J.; Dordick, J.S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020, 6, 50.
- Esposito, D.; Mehalko, J.; Drew, M.; Snead, K.; Wall, V.; Taylor, T.; Frank, P.; Denson, J.P.; Hong, M.; Gulten, G.; et al. Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein Expr. Purif. 2020, 174, 105686.
- Mouffak, S.; Shubbar, Q.; Saleh, E.; El-Awady, R. Recent advances in management of COVID-19: A review. Biomed. Pharmacother. 2021, 143, 112107.
- Monreal-Escalante, E.; Ramos-Vega, A.; Angulo, C.; Bañuelos-Hernández, B. Plant-Based Vaccines: Antigen Design, Diversity, and Strategies for High Level Production. Vaccines 2022, 10, 100.
- Venkataraman, S.; Hefferon, K.; Makhzoum, A.; Abouhaidar, M. Combating Human Viral Diseases: Will Plant-Based Vaccines Be the Answer? Vaccines 2021, 9, 761.
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.Y.; Couture, M.; D‘Aoust, M.A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078.
- Burnett, M.J.B.; Burnett, A.C. Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants People Planet 2020, 2, 121–132.
- Balieu, J.; Jung, J.-W.; Chan, P.; Lomonossoff, G.P.; Lerouge, P.; Bardor, M. Investigation of the N-Glycosylation of the SARS-CoV-2 S Protein Contained in VLPs Produced in Nicotiana benthamiana. Molecules 2022, 27, 5119.
- McGuire, B.E.; Mela, J.E.; Thompson, V.C.; Cucksey, L.R.; Stevens, C.E.; McWhinnie, R.L.; Winkler, D.F.H.; Pelech, S.; Nano, F.E. Escherichia coli recombinant expression of SARS-CoV-2 protein fragments. Microb. Cell Factories 2022, 21, 21.
- Argentinian AntiCovid Consortium. Structural and functional comparison of SARS-CoV-2-spike receptor binding domain produced in Pichia pastoris and mammalian cells. Sci. Rep. 2020, 10, 21779.
- Berndt, A.J.; Smalley, T.N.; Ren, B.; Simkovsky, R.; Badary, A.; Sproles, A.E.; Fields, F.J.; Torres-Tiji, Y.; Heredia, V.; Mayfield, S.P. Recombinant production of a functional SARS-CoV-2 spike receptor binding domain in the green algae Chlamydomonas reinhardtii. PLoS ONE 2021, 16, e0257089.
- Toustou, C.; Walet-Balieu, M.L.; Kiefer-Meyer, M.C.; Houdou, M.; Lerouge, P.; Foulquier, F.; Bardor, M. Towards understanding the extensive diversity of protein N-glycan structures in eukaryotes. Biol. Rev. Camb. Philos. Soc. 2022, 97, 732–748.
- Kiefer, A.; Niemeyer, J.; Probst, A.; Erkel, G.; Schroda, M. Production and secretion of functional full-length SARS-CoV-2 spike protein in Chlamydomonas reinhardtii. Front. Plant. Sci. 2021, 20, 988870.
- Malla, A.; Rosales-Mendoza, S.; Phoolcharoen, W.; Vimolmangkang, S. Efficient Transient Expression of Recombinant Proteins Using DNA Viral Vectors in Freshwater Microalgal Species. Front. Plant. Sci. 2021, 12, 650820.
- Siriwattananon, K.; Manopwisedjaroen, S.; Kanjanasirirat, P.; Budi Purwono, P.; Rattanapisit, K.; Shanmugaraj, B.; Smith, D.R.; Borwornpinyo, S.; Thitithanyanont, A.; Phoolcharoen, W. Development of Plant-Produced Recombinant ACE2-Fc Fusion Protein as a Potential Therapeutic Agent Against SARS-CoV-2. Front. Plant. Sci. 2020, 11, 604663.
- Xie, X.; Du, H.; Chen, J.; Aslam, M.; Wang, W.; Chen, W.; Li, P.; Du, H.; Liu, X. Global Profiling of N-Glycoproteins and N-Glycans in the Diatom Phaeodactylum tricornutum. Front. Plant Sci. 2021, 12, 779307.
- Baïet, B.; Burel, C.; Saint-Jean, B.; Louvet, R.; Menu-Bouaouiche, L.; Kiefer-Meyer, M.C.; Mathieu-Rivet, E.; Lefebvre, T.; Castel, H.; Carlier, A.; et al. N-glycans of Phaeodactylum tricornutum diatom and functional characterization of its N-acetylglucosaminyltransferase I enzyme. J. Biol. Chem. 2011, 286, 6152–6164.
- Dumontier, R.; Loutelier-Bourhis, C.; Walet-Balieu, M.L.; Burel, C.; Mareck, A.; Afonso, C.; Lerouge, P.; Bardor, M. Identification of N-glycan oligomannoside isomers in the diatom Phaeodactylum tricornutum. Carbohydr. Polym. 2021, 259, 117660.
