Time series classification (TSC) is very commonly used for modeling digital clinical measures. Time Series Classification (TSC) involves building predictive models that output a target variable or label from inputs of longitudinal or sequential observations across some time period. These inputs could be from a single variable or multiple variables measured across time, where the measurements can be ordinal or numerical (discrete or continuous).
- time series classification
- digital clinical measures
- machine learning
1. Introduction
Time series data are a very common form of data, containing information about the (changing) state of any variable. Some common examples include stock market prices and temperature values across some period of time. Time series modeling tasks include classification, regression, and forecasting. There are unique challenges that come with modeling time series, given that measurements obtained in real-life settings are subject to random noise, and that any measurement at a particular point in time could be related to or influenced by measurements at other points in time [1]. Given this nature of time series data, it is impractical to simply utilize established machine learning algorithms such as logistic regression, support vector machine, or random forest on the raw time series datasets because these data violate the basic assumptions of those models. In recent years, two vastly different camps of time series classification techniques have emerged: deep-learning-based models vs non-deep-learning-based models. While deep learning models are extremely powerful and show great promise in classification performance and generalizability, they also present challenges in the areas of hyperparameter tuning, training, and model complexity decisions.
2. Time Series Classification Techniques Used in Biomedical Applications
2.1. Preprocessing Methods
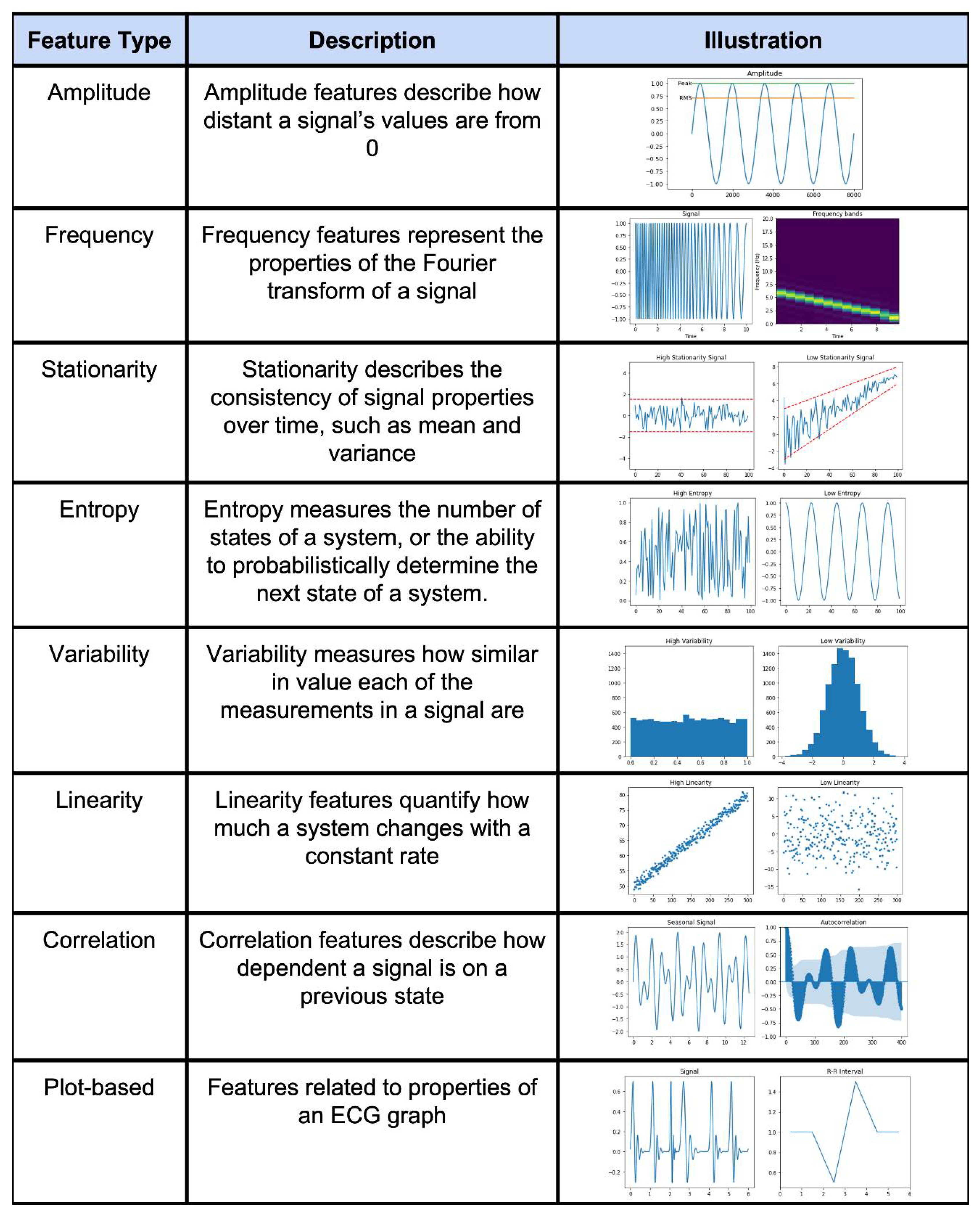
2.2. Feature Engineering Methods
-
Preprocessing: this step takes raw data as the input and performs some manipulation of the data to return cleaner signals. Common steps include artifact removal, filtering, and segmentation.
-
Signal transformation: this step can be used in preprocessing and also as a precursor to feature extraction. Some manipulation is performed on the signal to represent it in a different space. Common choices are Fourier Transform and wavelet transforms.
-
Feature extraction: in this step, features are extracted from the time series data as a new representation of the original time series.
-
Feature selection: this step selects the features that are the most descriptive, or have the most explanation power. Feature selection is also frequently performed in conjunction with model building.
-
Model selection: the best model is found through hyperparameter tuning and/or comparisons between different types of algorithms.
-
Model validation: performance metrics are calculated for all of the final models. This is frequently done in conjunction with model selection and often using some form of cross-validation.
2.3. Other Methods
2.4. Interpretation Methods
2.5. Best Performing Algorithms
Overall, the statistical modeling classifiers and feature engineering methods performed the best and most consistently for all input signal types. Wavelet transformation is consistently and widely used and achieving great performances as a preprocessing method, feature extraction method, or as an integral part of index development.
3. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/s22208016
References
- Bock, C.; Moor, M.; Jutzeler, C.R.; Borgwardt, K. Machine Learning for Biomedical Time Series Classification: From Shapelets to Deep Learning. In Artificial Neural Networks; Cartwright, H., Ed.; Springer: New York, NY, USA, 2021; pp. 33–71.
- Mole, S.S.S.; Sujatha, K. An efficient Gait Dynamics classification method for Neurodegenerative Diseases using Brain signals. J. Med. Syst. 2019, 43, 245.
- Joshi, D.; Khajuria, A.; Joshi, P. An automatic non-invasive method for Parkinson’s disease classification. Comput. Methods Programs Biomed. 2017, 145, 135–145.
- Tor, H.T.; Ooi, C.P.; Lim-Ashworth, N.S.; Wei, J.K.E.; Jahmunah, V.; Oh, S.L.; Acharya, U.R.; Fung, D.S.S. Automated detection of conduct disorder and attention deficit hyperactivity disorder using decomposition and nonlinear techniques with EEG signals. Comput. Methods Programs Biomed. 2021, 200, 105941.
- Mesbah, S.; Gonnelli, F.; Angeli, C.A.; El-Baz, A.; Harkema, S.J.; Rejc, E. Neurophysiological markers predicting recovery of standing in humans with chronic motor complete spinal cord injury. Sci. Rep. 2019, 9, 14474.
- Anh, N.X.; Nataraja, R.; Chauhan, S. Towards near real-time assessment of surgical skills: A comparison of feature extraction techniques. Comput. Methods Progr. Biomed. 2019, 187, 105234.
- Durongbhan, P.; Zhao, Y.; Chen, L.; Zis, P.; De Marco, M.; Unwin, Z.C.; Venneri, A.; He, X.; Li, S.; Zhao, Y.; et al. A Dementia Classification Framework Using Frequency and Time-Frequency Features Based on EEG Signals. IEEE Trans. Neural Syst. Rehabil. Eng. Publ. IEEE Eng. Med. Biol. Soc. 2019, 27, 826–835.
- Bhattacharya, S.; Mazumder, O.; Roy, D.; Sinha, A.; Ghose, A. Synthetic Data Generation Through Statistical Explosion: Improving Classification Accuracy of Coronary Artery Disease Using PPG. In Proceedings of the ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1165–1169.
- Walter, S.; Gruss, S.; Limbrecht-Ecklundt, K.; Traue, H.C.; Werner, P.; Al-Hamadi, A.; Diniz, N.; da Silva, G.M.; Andrade, A.O. Automatic pain quantification using autonomic parameters. Psychol. Neurosci. 2014, 7, 363–380.
- Ruiz, A.P.; Flynn, M.; Large, J.; Middlehurst, M.; Bagnall, A. The great multivariate time series classification bake off: A review and experimental evaluation of recent algorithmic advances. Data Min. Knowl. Discov. 2020, 35, 401–449.
- She, X.; Zhai, Y.; Henao, R.; Woods, C.W.; Chiu, C.; Ginsburg, G.S.; Song, P.X.K.; Hero, A.O. Adaptive Multi-Channel Event Segmentation and Feature Extraction for Monitoring Health Outcomes. IEEE Trans. Biomed. Eng. 2020, 68, 2377–2388.
- Zhou, P.-Y.; Chan, K.C.C. Fuzzy Feature Extraction for Multichannel EEG Classification. IEEE Trans. Cogn. Dev. Syst. 2016, 10, 267–279.
- Forestier, G.; Petitjean, F.; Riffaud, L.; Jannin, P. Automatic matching of surgeries to predict surgeons’ next actions. Artif. Intell. Med. 2017, 81, 3–11.
- Iscan, M.; Yigit, F.; Yilmaz, C. Heartbeat pattern classification algorithm based on Gaussian mixture model. In Proceedings of the 2016 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Benevento, Italy, 15–18 May 2016; pp. 1–6.
- Zhou, H.; Ji, N.; Samuel, O.W.; Cao, Y.; Zhao, Z.; Chen, S.; Li, G. Towards Real-Time Detection of Gait Events on Different Terrains Using Time-Frequency Analysis and Peak Heuristics Algorithm. Sensors 2016, 16, 1634.
