Complex organisms are associations of different cells that coexist and collaborate creating a living consortium, the holobiont. The relationships between the holobiont members are essential for a proper homeostasis of all the organisms and they are founded on the establishment of complex inter-connections between all the cells. Non-coding RNAs are transcriptional products of the genomic output with regulatory function and they can act as communication signals between cells, being involved either in homeostasis or dysbiosis of the holobiont. Eukaryotic and prokaryotic cells can transmit signals via non-coding RNAs using specific extracellular conveyors that will travel to reach the target cell and will be translated into a regulatory response by a dedicated molecular machinery. Within holobionts, non-coding RNA regulatory signaling is involved in symbiotic and pathogenic relationships among cells. This review analyzes the current knowledge about the role of non-coding RNAs in cell-to-cell communication, with a special focus in the signaling between cells in multi-organism consortia.
- ncRNA
- holobiont
- communication
1. Diversity of RNA Species in Transcriptomic Output
In 1968, Francis Crick published a paper describing the role of the dynamics of genetic information in the evolution and the fundaments of the origins of the genetic code in living cells. This paper is considered to be the embryonic core of the “RNA world” theory[1]. The RNA world stands on the idea that RNA was the first “living molecule”, being in the origin of the evolution of all biological macromolecules and, consequently, acting as a biological platform from which other more complex biomolecules will arise[2]. RNA was favored in pre-biotic molecular evolution due to its intrinsic versatility and adaptability, and because it can carry sequence and structure information. In consequence RNA must be placed at the root of the molecular tree of life[3]. In most cases proteins have become more efficient to perform the same catalytic tasks, and they have been delegated to those functions along evolution, despite the existence of catalytic RNA molecules, as represented by ribozymes and the rRNAs. This fact denotes a clear Darwinian pattern of molecular evolution events, where a function is acquired and further improved and radiated into new ones by environmental pressure[3].
The dynamics of genomic output either in eukaryotic or in prokaryotic cells is largely determined by the pervasive transcription phenomenon[4][5]. Genome-wide transcription in all living cells is responsible for the biosynthesis of two main classes of RNA molecules: The coding RNAs will be translated to produce proteins, and the non-coding RNAs (ncRNAs) constituted by transcripts with reduced coding potential will not be translated[6]. ncRNAs are regulatory molecules that are generated either from specific transcriptional units or by the processing of already existing RNAs with alternative functions. In complex eukaryotic organisms, ncRNAs comprise most of the transcriptome, which suggests the importance of this second evolutionary tier of genetic output that might enable the integration and coordination of sophisticated suites of gene expression required for differentiation and development[7].
The non-coding transcriptome of eukaryotic cells includes several families of molecular species that are typically classified by their size, biogenesis pathways, and regulatory functions. The most widely used ncRNA classification criteria divides them in two families: short ncRNAs, comprised by molecules less than 200 nt in length, and long ncRNAs, a very diverse group of high-molecular weight ncRNAs, with more than 200 nt[8]. The short ncRNAs include the well-known groups of ncRNA involved in protein translation (the 5.8S and 5S rRNAs, and tRNAs) and RNA splicing and modification (snoRNAs), together with other regulatory ncRNAs, such as microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), and small interfering RNAs (siRNAs)[9]. miRNAs are one of the most versatile families of small eukaryotic ncRNAs composed by single-stranded short RNA molecules (19–23 nt) generated from the transcription and further processing of selected transcriptional units that act as negative regulators of the genomic output at the post-transcriptional level[10] . Ineukaryotic cells, the family of long ncRNAs can be also divided in two subgroups: long non-coding RNAs (lncRNAs), transcriptional products of genomic loci with very limited coding potential that can regulate the genomic output at many levels from transcription to translation[11], and circular RNAs (circRNAs), covalently closed ncRNAs that result from non-canonical back splicing events of coding or non-coding transcriptional units that typically act as molecular sponges interacting with proteins or other ncRNAs[12]. The abundance and expansion of the repertoire of ncRNAs in higher eukaryotic organisms characterize them as a molecular players of evolution, because the organisms could not be dependent on a limited number of proteins or protein isoforms, but on a much larger set of genomic instructions that are embedded in trans-acting RNAs[13]. It is also very likely that dynamic ncRNA-regulatory networks can contribute much of the phenotypic variation that is observed between individuals and species and it can be involved in the response to internal and external stimuli[10].
In prokaryotic organisms, genome size and structure limit the heterogeneity of the transcriptional output when compared with eukaryotic cells. However, several ncRNA families have been characterized as relevant players in bacterial physiology and homeostasis. For instance, small bacterial ncRNA (sRNAs) are typically 50 to 400 nucleotides in length and, despite their lower abundance and diversity when compared with eukaryotic cells, they are important regulators of gene expression in specific conditions, like stress response, quorum sensing, and virulence[14][15]. Coleman and coworkers described the first evidence of a functional sRNA of bacterial origin with the characterization of micF, a non-coding RNA complementary to the Shine-Dalgarno sequence of the E. coli lpp mRNA that was able to silence its expression[16]. Bacterial sRNAs belong to two functional families: the cisRNAs originated by antisense transcription of coding genes that act over the parental coding frames by complementary base pairing and they are generally involved in processes, such as the maintenance of the copy number in plasmids or the regulation of operons[17] and the trans-encoded sRNAs that are generated by specific transcriptional units and involved in the post-transcriptional regulation of mRNAs[18]. Moreover, the recent developments of next-generation sequencing approaches for transcriptomic analysis allowed for the characterization of new populations of bacterial sRNAs derived from the post-transcriptional processing of other RNAs[13]. Because of their smaller size (16–25 nt), this family of bacterial sRNAs is often referred as “miRNA-like” and they have been characterized as co-adjuvant virulence factors in some pathogenic bacteria[19].
The importance of RNA molecules makes RNA an ideal player for the interchange of information between cells. There is a common ancient language encoded in RNA molecules, independent of their origins, which can be understood by cells and translated into metabolic actions. The putative roles of an RNA-based language are particularly interesting when the relationships between different species are analyzed. The role of RNA molecules, more specifically ncRNAs, in the interchange of information between cells is starting to be unveiled with the characterization of RNA-based communication mechanisms in binary systems, but more knowledge is required in order to understand the detailed functions of ncRNA regulatory networks in the context of holobionts[20][21].
2. Conveyors for ncRNA Transport between Cells
Valadi and coworkers observed RNA transfer between cells under physiological conditions and without involving the destruction of the cell structure for the first time in mouse and human cell lines in 2007[22]. This functional transfer was mediated by extracellular vesicles, namely exosomes, containing diverse coding and non-coding RNAs. Exosome-mediated RNA transfer from mouse into human cells was sufficient to release coding RNA molecules that were translated to human proteins in the mouse cell line, but the exosomes were also loaded with numerous miRNAs that could have functional effects in the recipient cell[23]. Functional ncRNA transfer between tissues at the organism level was further demonstrated in animal models, showing the specific release of exosome-encapsulated miRNAs from niches, such as the adipose tissue, their transport across the animal body by circulating fluids, and their final regulatory action over distant cells[20].
Extracellular vesicles (EVs), which are the major conveyors of ncRNAs in their cell-to-cell journey, can be classified according to their diameter and their biogenesis pathways[21]. EVs that are generated by the exocytic pathway in eukaryotic cells are designated as exosomes and are 30–150 nm in diameter. Ectosomes or shedding microvesicles are generated by the outward budding of cell membranes and they are typically larger than exosomes with a diameter between 100–1000 nm. Later stages of apoptosis are responsible for the generation of a third family of EVs, the apoptotic bodies, which are a very heterogeneous family of vesicles that present diameters that ranged from 50–5000 nm[22]. Bacteria are also able to generate nano-sized EVs either constitutively or as a response to environmental stimuli[24]. EVs of bacterial origin were initially described in Gram negative microorganisms as the result of the shedding of the outer-membrane and they were consequently designated as outer membrane vesicles (OMV). Recent evidence also showed that Gram positive bacteria can produce EVs, but the mechanism for the biogenesis and release of those microvesicles are still not completely characterized[13][25].
Independent of their biological origin, there are still many open issues regarding the functions of EVs as carriers of ncRNAs, namely those that were related with the mechanisms used by the cells to select and process ncRNAs as vesicle cargo (Figure 1). The current accumulated knowledge about the mechanisms of ncRNAs sorting into secretory vesicles is focused on miRNAs in human and mammalian cells. Experimental evidence showed that the miRNA sorting into exosomes and other vesicles is mediated by RNA-binding and cytoskeleton proteins, following patterns that appear to be cell specific[26][27][28]. Interestingly, in human cells, the external perturbation of the expression of individual miRNAs or their cognate targets promotes bidirectional miRNA relocation from the cytoplasm to multivesicular bodies, controlling the miRNA sorting to exosomes[29]. The mechanisms of sorting ncRNA into vesicles remain unknown in lower eukaryotes and bacteria.
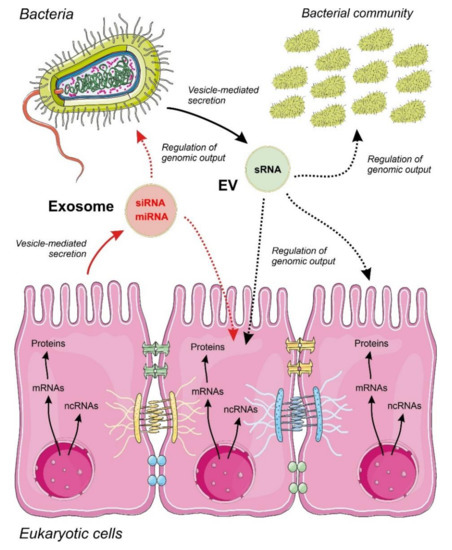
Figure 1. Proposed model for inter-regulatory networks mediated by non-coding RNAs (ncRNAs) secreted within membrane-containing vesicles in a binary system composed by a eukaryotic epithelium and an associated bacterial community. Small regulatory ncRNAs produced by eukaryotic cells (miRNAs and siRNAs) and bacteria (sRNAs) are selected, sorted and secreted by vesicles acting as regulators of the genomic output crossing cell boundaries. The specificity and detailed mechanisms involved in ncRNA selection and sorting by the cells are largely unknown. The widespread regulatory language transported and exerted by ncRNA molecules allow a cross-kingdom communication and an efficient interaction between different cell types. Metabolic flow from genomic information is depicted by continuous arrows and regulatory events controlled by ncRNAs by dotted arrows.
3. Molecular Machinery for Understanding the External ncRNA Message
Once the extracellular signals that were transported by ncRNA reach the target cell, they need to be translated into functional actions to generate a physiological response. Within the holobiont context, the ncRNA-mediated signals need to be efficiently understood by the target cell and ideally by different target cells. The efficient communication by ncRNAs is ensured by the plasticity of RNA molecules, their presence in all living cells, and shared mechanisms of action among distinct organisms. The small RNAs produced by eukaryotic (miRNAs and siRNAs)[30][31] and prokaryotic cells (sRNAs) illustrate the inter-cell communication that is mediated by ncRNAs[32][33]. The regulatory roles of other ncRNA families in cross-species communication, such as circRNAs or lncRNAs, remain unknown.
In eukaryotic cells, external small RNAs that are transported by EVs are internalized either by vesicle endocytosis or by membrane fusion and posterior release of vesicle cargo into the cytoplasm. The mechanisms of a hypothetical targeting of ncRNA-containing vesicles to specific cells remain unknown. Signals that are transported by small ncRNAs (miRNAs, siRNAs or bacterial sRNAs) and targeting eukaryotic cells are expected to function by the insertion of these external molecules into the canonical regulatory pathways that are mediated by ncRNAs in the cell. Experimental evidence suggests that foreign regulatory small ncRNAs would function by base pairing to their molecular targets, which are usually mRNAs as well as ncRNAs[34]. It is important to note that the regulatory action of any small ncRNA is strictly dependent on the presence of the corresponding cognate target in the recipient cell. The same small ncRNA could have different regulatory effects, depending not only on the targeted cell, but also on its physiological state. Moreover, small ncRNAs would also need the help of specific RNA binding proteins that will be responsible for the final regulatory action.
In eukaryotes, small ncRNAs are recognized by conserved RNA-binding proteins that belong to the argonaute family (Ago)[35]. Canonical and Ago-like proteins are encoded by the vast majority of eukaryotic genomes from fungi[36][37] to plants[38][39] and animals[40], ensuring a molecular background for the action of internal or external small ncRNAs[41][42]. In the cytoplasm of animal cells, a member of the argonaute protein family Ago2 is a key component of the RNA-induced silencing complex (RISC), which the base complementarity between the small RNA and its target will guide. If the complementarity between the small ncRNA and the target is close to 100%, then the endonuclease activity of Ago2 will be responsible for the targeted cleavage of the target transcript that will be further degraded by RNases. In the case of partial complementarity, the canonical miRNA pathway will induce a RISC-mediated translational repression by a mechanism that often involves transcript deadenylation and further decay[10][43]. In plants, Ago1 and Ago2 proteins are generally involved in sRNA-mediated gene silencing, which occurs by target RNA degradation due to the high degree of complementarity between the RNA molecules[44]. Solid experimental evidence supported the function of externally-originated small RNAs in order to regulate gene expression within different eukaryotic hosts[45][46].
Bacteria also have regulatory networks that are centered on small ncRNA molecules. The first evidence for the characterization of the protein players of these RNA regulatory networks pointed to the importance of the Hfq protein, a small RNA chaperone that was detected in association with small ncRNAs and mRNA transcripts[47][48]. Further experimental data described the additional existence of ribonucleoprotein complexes that were integrated by the Hfq chaperone together with RNase E that were responsible for the post-transcriptional silencing and degradation of target mRNAs by the action of specific families of complementary sRNAs[49][50]. Hfq-dependent RNA regulatory networks have been involved in virulence and pathogenesis processes in several bacteria[51][52], but they are also expected to be involved in the regulatory action of exogenously acquired ncRNAs[30][53]. In contrast to eukaryotic cells, the effect of bacterial ncRNAs over mRNA transcripts is different from canonical miRNA/siRNA regulation and it could also include an enhancement in the stability and expression of the targeted transcripts[30][53].
This entry is adapted from the peer-reviewed paper 10.3390/ijms21072333
References
- F.H.C. Crick; The origin of the genetic code. Journal of Molecular Biology 1968, 38, 367-379, 10.1016/0022-2836(68)90392-6.
- T. Cech; RNA world. Nature 1987, 328, 676-676, 10.1038/328676b0.
- Irma Lozada-Chávez; Peter F. Stadler; Sonja J. Prohaska; “Hypothesis for the Modern RNA World”: A pervasive Non-coding RNA-Based Genetic Regulation is a Prerequisite for the Emergence of Multicellular Complexity. Origins of Life and Evolution of Biospheres 2011, 41, 587-607, 10.1007/s11084-011-9262-1.
- Marcel E Dinger; Paulo De Paiva Amaral; Timothy R. Mercer; John Mattick; Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Briefings in Functional Genomics and Proteomics 2009, 8, 407-423, 10.1093/bfgp/elp038.
- Matthew J. Hangauer; Ian W. Vaughn; Michael T. McManus; Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLOS Genetics 2013, 9, e1003569, 10.1371/journal.pgen.1003569.
- Philipp Kapranov; Jill Cheng; Sujit Dike; David A. Nix; Radharani Duttagupta; Aarron T. Willingham; Peter F. Stadler; Jana Hertel; Jörg Hackermüller; Ivo Hofacker; et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484-1488, 10.1126/science.1138341.
- Sonja Hombach; Markus Kretz; Non-coding RNAs: Classification, Biology and Functioning. Advances in Experimental Medicine and Biology 2016, 937, 3-17, 10.1007/978-3-319-42059-2_1.
- John Mattick; Igor V Makunin; Small regulatory RNAs in mammals. Human Molecular Genetics 2005, 14, R121–R132, 10.1093/hmg/ddi101.
- Sergej Djuranovic; Ali Nahvi; Rachel Green; miRNA-Mediated Gene Silencing by Translational Repression Followed by mRNA Deadenylation and Decay. Science 2012, 336, 237-240, 10.1126/science.1215691.
- Ioannis Sarropoulos; Ray Marin; Margarida Cardoso-Moreira; Henrik Kaessmann; Developmental dynamics of lncRNAs across mammalian organs and species.. Nature 2019, 571, 510-514, 10.1038/s41586-019-1341-x.
- Chuan Huang; Ge Shan; What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription 2015, 6, 61-64, 10.1080/21541264.2015.1071301.
- Alex C. Tuck; Kedar Nath Natarajan; Greggory Rice; Jason Borawski; Fabio Mohn; Aneliya Rankova; Matyas Flemr; Alice Wenger; Razvan Nutiu; Sarah A. Teichmann; et al. Distinctive features of lincRNA gene expression suggest widespread RNA-independent functions. Life Science Alliance 2018, 1, e201800124, 10.26508/lsa.201800124.
- Marie-Claude Carrier; David Lalaouna; Eric Massé; Broadening the Definition of Bacterial Small RNAs: Characteristics and Mechanisms of Action. Annual Review of Microbiology 2018, 72, 141-161, 10.1146/annurev-micro-090817-062607.
- Marie-Claude Carrier; David Lalaouna; Eric Massé; Broadening the Definition of Bacterial Small RNAs: Characteristics and Mechanisms of Action. Annual Review of Microbiology 2018, 72, 141-161, 10.1146/annurev-micro-090817-062607.
- Jack Coleman; Pamela J. Green; Masayori Inouye; The use of RNAs complementary to specific mRNAs to regulate the expression of individual bacterial genes. Cell 1984, 37, 429-436, 10.1016/0092-8674(84)90373-8.
- Jason A. Opdyke; Elizabeth M. Fozo; Matthew R. Hemm; Gisela Storz; RNase III Participates in GadY-Dependent Cleavage of the gadX-gadW mRNA. Journal of Molecular Biology 2010, 406, 29-43, 10.1016/j.jmb.2010.12.009.
- Yuhua Zhan; Zhiping Deng; Yongliang Yan; Hongyang Zhang; Chao Lu; Zhimin Yang; Liguo Shang; Yi Huang; Fanyang Lv; Yaqun Liu; et al. NfiR, a New Regulatory Noncoding RNA (ncRNA), Is Required in Concert with the NfiS ncRNA for Optimal Expression of Nitrogenase Genes in Pseudomonas stutzeri A1501.. Applied and Environmental Microbiology 2019, 85, e00762-19, 10.1128/AEM.00762-19.
- J.-W. Choi; S.-C. Kim; S.-H. Hong; H.-J. Lee; Secretable Small RNAs via Outer Membrane Vesicles in Periodontal Pathogens. Journal of Dental Research 2017, 96, 458-466, 10.1177/0022034516685071.
- Jean‐Christophe Simon; Julian R. Marchesi; Christophe Mougel; Marc-André Selosse; Host-microbiota interactions: from holobiont theory to analysis. Microbiome 2019, 7, 5, 10.1186/s40168-019-0619-4.
- Thomas Thomou; Marcelo A. Mori; Jonathan M. Dreyfuss; Masahiro Konishi; Masaji Sakaguchi; Christian Wolfrum; Tata Nageswara Rao; Jonathon N. Winnay; Ruben Garcia-Martin; Steven K. Grinspoon; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450-455, 10.1038/nature21365.
- Leonid Margolis; Yoel Sadovsky; The biology of extracellular vesicles: The known unknowns.. PLoS Biology 2019, 17, e3000363, 10.1371/journal.pbio.3000363.
- Hina Kalra; Gregor Drummen; Suresh Mathivanan; Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. International Journal of Molecular Sciences 2016, 17, 170, 10.3390/ijms17020170.
- Hadi Valadi; Karin Ekström; Apostolos Bossios; Margareta Sjöstrand; James J Lee; Jan Lötvall; Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature 2007, 9, 654-659, 10.1038/ncb1596.
- Ji Hyun Kim; Jaewook Lee; Jaesung Park; Yong Song Gho; Gram-negative and Gram-positive bacterial extracellular vesicles. Seminars in Cell & Developmental Biology 2015, 40, 97-104, 10.1016/j.semcdb.2015.02.006.
- Sara Ahmadi Badi; Arfa Moshiri; Abolfazl Fateh; Fatemeh Rahimi Jamnani; Meysam Sarshar; Farzam Vaziri; Seyed Davar Siadat; Microbiota-Derived Extracellular Vesicles as New Systemic Regulators. Frontiers in Microbiology 2017, 8, 1610, 10.3389/fmicb.2017.01610.
- Heedoo Lee; Chunhua Li; Yang Zhang; Duo Zhang; L. E. Otterbein; Yang Jin; Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. Journal of Experimental Medicine 2019, 216, 2202-2220, 10.1084/jem.20182313.
- Morayma Temoche-Diaz; Matthew Shurtleff; Ryan Nottingham; Jun Yao; Raj P. Fadadu; Alan M. Lambowitz; Randy Schekman; Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife 2019, 8, 612069, 10.1101/612069.
- Matthew Shurtleff; Kate V. Karfilis; Morayma Temoche-Diaz; Sayaka Ri; Randy Schekman; Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 2016, 5, e19276, 10.1101/040238.
- Mario Leonardo Squadrito; Caroline Baer; Frederic Burdet; Claudio Maderna; Gregor D. Gilfillan; Robert Lyle; Mark Ibberson; Michele De Palma; Endogenous RNAs Modulate MicroRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cell Reports 2014, 8, 1432-1446, 10.1016/j.celrep.2014.07.035.
- Shirong Liu; Andre Pires Da Cunha; Rafael M. Rezende; Ron Cialic; Zhiyun Wei; Lynn Bry; Laurie E. Comstock; Roopali Gandhi; Howard L. Weiner; The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host & Microbe 2016, 19, 32-43, 10.1016/j.chom.2015.12.005.
- Emilie Viennois; Benoit Chassaing; Anika Tahsin; Adani Pujada; Lixin Wang; Andrew T. Gewirtz; Didier Merlin; Host-derived fecal microRNAs can indicate gut microbiota healthiness and ability to induce inflammation.. Theranostics 2019, 9, 4542-4557, 10.7150/thno.35282.
- Cherie Blenkiron; Denis Simonov; Anitadevi Muthukaruppan; Peter Tsai; Priscila Dauros; Sasha Green; Jiwon Hong; Cristin Print; Simon Swift; Anthony R. Phillips; et al. Uropathogenic Escherichia coli Releases Extracellular Vesicles That Are Associated with RNA. PLOS ONE 2016, 11, e0160440, 10.1371/journal.pone.0160440.
- Katja Koeppen; Thomas H. Hampton; Michael Jarek; Maren Scharfe; Scott A. Gerber; Daniel W. Mielcarz; Elora G. Demers; Emily L. Dolben; John H. Hammond; Deborah A. Hogan; et al. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLOS Pathogens 2016, 12, e1005672, 10.1371/journal.ppat.1005672.
- Ming Wang; Arne Weiberg; Feng-Mao Lin; Bart P. H. J. Thomma; Hsien-Da Huang; Hailing Jin; Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nature Plants 2016, 2, 16151-16151, 10.1038/nplants.2016.151.
- Michelle A. Carmell; Zhenyu Xuan; Michael Q. Zhang; Gregory J. Hannon; The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Development 2002, 16, 2733-2742, 10.1101/gad.1026102.
- Laurence Braun; Dominique Cannella; Philippe Ortet; Mohamed Barakat; Céline F. Sautel; Sylvie Kieffer; Jérome Garin; Olivier Bastien; Olivier Voinnet; Mohamed-Ali Hakimi; et al. A Complex Small RNA Repertoire Is Generated by a Plant/Fungal-Like Machinery and Effected by a Metazoan-Like Argonaute in the Single-Cell Human Parasite Toxoplasma gondii. PLOS Pathogens 2010, 6, e1000920, 10.1371/journal.ppat.1000920.
- Quyet Nguyen; Akihide Iritani; Shuhei Ohkita; Ba V Vu; Kana Yokoya; Ai Matsubara; Ken-Ichi Ikeda; Nobuhiro Suzuki; Hitoshi Nakayashiki; A fungal Argonaute interferes with RNA interference.. Nucleic Acids Research 2018, 46, 2698-2698, 10.1093/nar/gky078.
- Károly Fátyol; Márta Ludman; József Burgyán; Functional dissection of a plant Argonaute.. Nucleic Acids Research 2015, 44, 1384-97, 10.1093/nar/gkv1371.
- Han Zhang; Rui Xia; Blake C. Meyers; Virginia Walbot; Evolution, functions, and mysteries of plant ARGONAUTE proteins. Current Opinion in Plant Biology 2015, 27, 84-90, 10.1016/j.pbi.2015.06.011.
- Takashi Sasaki; Aiko Shiohama; Shinsei Minoshima; Nobuyoshi Shimizu; Identification of eight members of the Argonaute family in the human genome☆. Genomics 2003, 82, 323-330, 10.1016/s0888-7543(03)00129-0.
- Julia Hock; Gunter Meister; The Argonaute protein family. Genome Biology 2008, 9, 210-210, 10.1186/gb-2008-9-2-210.
- Xiang Zhou; Heng Guo; Ke Chen; Hanhua Cheng; Rongjia Zhou; Identification, chromosomal mapping and conserved synteny of porcine Argonaute family of genes. Genetica 2010, 138, 805-812, 10.1007/s10709-010-9462-z.
- Ana Eulalio; Eric Huntzinger; Tadashi Nishihara; Jan Rehwinkel; Maria Fauser; Elisa Izaurralde; Deadenylation is a widespread effect of miRNA regulation. RNA 2008, 15, 21-32, 10.1261/rna.1399509.
- Chien-Yu Huang; Huan Wang; Po Hu; Rachael Hamby; Hailing Jin; Small RNAs - Big Players in Plant-Microbe Interactions.. Cell Host & Microbe 2019, 26, 173-182, 10.1016/j.chom.2019.07.021.
- Amy H Buck; Gillian Coakley; Fabio Simbari; Henry J. McSorley; Juan F. Quintana; Thierry Le Bihan; Sujai Kumar; Cei Abreu-Goodger; Marissa Lear; Yvonne Harcus; et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nature Communications 2014, 5, 5488, 10.1038/ncomms6488.
- Saima Shahid; Gunjune Kim; Nathan Johnson; Eric Wafula; Feng Wang; Ceyda Coruh; Vivian Bernal-Galeano; Tamia Phifer; Claude W. Depamphilis; James H. Westwood; et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 2018, 553, 82-85, 10.1038/nature25027.
- Aixia Zhang; Karen M. Wassarman; Carsten Rosenow; Brian C. Tjaden; Gisela Storz; Susan Gottesman; Global analysis of small RNA and mRNA targets of Hfq. Molecular Microbiology 2003, 50, 1111-1124, 10.1046/j.1365-2958.2003.03734.x.
- Kimika Maki; Kanako Uno; Teppei Morita; Hiroji Aiba; RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proceedings of the National Academy of Sciences 2008, 105, 10332-10337, 10.1073/pnas.0803106105.
- Hiroji Aiba; Mechanism of RNA silencing by Hfq-binding small RNAs. Current Opinion in Microbiology 2007, 10, 134-139, 10.1016/j.mib.2007.03.010.
- Nicholas De Lay; Daniel J. Schu; Susan Gottesman; Bacterial Small RNA-based Negative Regulation: Hfq and Its Accomplices*. Journal of Biological Chemistry 2013, 288, 7996-8003, 10.1074/jbc.R112.441386.
- Giulia Oliva; Tobias Sahr; Monica Rolando; Maike Knoth; Carmen Buchrieser; A Unique cis-Encoded Small Noncoding RNA Is Regulating Legionella pneumophila Hfq Expression in a Life Cycle-Dependent Manner. mBio 2017, 8, e02182-16, 10.1128/mBio.02182-16.
- Chelsea A. Schiano; Lauren E. Bellows; Wyndham Lathem; The Small RNA Chaperone Hfq Is Required for the Virulence of Yersinia pseudotuberculosis. Infection and Immunity 2010, 78, 2034-2044, 10.1128/iai.01046-09.
- Mirza Abdul Qayyum; Saif Ul Islam; Muhammad Qasim; Liande Wang; Host-Pathogen interactions modulated by small RNAs. RNA Biology 2017, 14, 891-904, 10.1080/15476286.2017.1318009.
