Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was first detected in China in a group of patients suffering from pneumonia. It was established as the causative agent for COVID-19, the virus that led to the ongoing pandemic, and the recently raised threat of long COVID-19. Even though some cases can progress to life-threatening pneumonia, most individuals infected with the virus suffer from mild to moderate illness.
- SARS-CoV-2-on-chip
- long COVID
- RT-PCR
1. Developments in SARS-CoV-2 Diagnostic Strategies
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was first detected in China in a group of patients suffering from pneumonia. It was established as the causative agent for COVID-19, the virus that led to the ongoing pandemic, and the recently raised threat of long COVID-19. Even though some cases can progress to life-threatening pneumonia, most individuals infected with the virus suffer from mild to moderate illness. The various components in the SARS-CoV-2 virus are structural proteins, namely, (i) spike glycoprotein (S), (ii) nucleocapsid protein (N), (iii) matrix protein (M), and (iv) envelope protein (E), five to eight accessory proteins, ribonucleic acid (RNA), and sixteen non-structural proteins [1]. The spike protein helps the virus to attach, fuse, enter, and transmit into the human body. Therefore, it is important to ensure rapid and sensitive detection and diagnosis to provide antiviral treatment and control the spread of infection. As a large number of infected people tend to be asymptomatic, the virus can spread to other people while the initial host will remain free of any symptoms, hence showing a need for testing availability.
The most widely used method to detect SARS-CoV-2 virus infection nowadays is the reverse transcription polymerase chain reaction (RT-PCR) test. However, other methods, such as computerized tomography (CT) imaging and whole genome sequencing have been used to diagnose infected individuals [2]. In addition, the enzyme-linked immunosorbent assay (ELISA) and lateral flow immunoassay (LFIA) are also viable methods for COVID-19 diagnosis. However, the major drawbacks associated with these diagnosis methods are that they are time-intensive and require trained technicians and the use of expensive laboratory equipment. This necessitates the development of point-of-care biosensors for rapid testing and the quicker identification of infected patients [3]. Biosensors make use of the physiochemical detection of chemical substances containing a biological component. The use of point-of-care biosensors can decrease the time taken for an analysis to a few minutes instead of the several hours required in conventional methods, allowing patients to receive healthcare sooner and contain the spread of the virus [4].
Point-of-care testing helps to develop novel chip-based and paper-based biosensors for the fast and low-cost diagnosis of viral diseases [6,7]. These devices help to detect antigens, antibodies, and nucleic acids in saliva, blood, and phlegm. Point-of-care biosensors test these samples based on fluorescent, colorimetric, or electrochemical detection techniques [8,9]. Such devices offer many advantages compared to the other techniques used for detection, such as higher sensitivity, lower cost, high specificity, less time-intensive process, and greater user-friendliness. Additionally, because the results can be retrieved quickly and easily, they enable prompt detection, which lowers the danger of transmission.
2. Understanding Long COVID: Cause, Symptoms, and Detection Fundamentals
This section covers the structure and biomarkers of the long COVID causative virus and explains the symptoms faced by an individual infected by the SARS-CoV-2 virus. It also discusses how the virus can be detected by its RNA or through antigen–antibody interactions.
2.1. Structure and Biomarkers of SARS-CoV-2 Virus
Similar to the other known coronaviruses, SARS-CoV-2 is an enveloped, single-stranded RNA virus [11]. It has a genome consisting of positive-sense single-stranded RNAs and nucleotides with 30,000 bases [12,13,14]. Apart from the four proteins responsible for the structural integrity of the SARS-CoV-2 virus, as discussed previously, about 27 different proteins are encoded in the viral genome, which comprises an RNA-dependent RNA-polymerase (RdRP). This RdRP interacts with the other non-structural proteins and plays a vital role in maintaining the fidelity of the genomic material [13,15,16]. SARS-CoV-2 virus binding with the host cell receptor is assisted by the surface spike glycoprotein gene encoded by coronaviruses; this is attributed to the receptor-binding domain (RBD) of the surface spike protein [17]. The surface spike protein was found to mediate the interaction with the help of angiotensin-converting enzyme 2 (ACE2), membrane fusion, a host receptor, and viral entry [18]. It has also been found to be responsible for the basic reproduction number and the determination of host tropism [18]. Less than 75% of the nucleotide sequence of the spike gene is similar to that of other members of the SARS coronaviruses. The remaining nucleotide sequence can be variable, thus making the spike gene quite diverse [19]. Other than this spike protein, the other three proteins responsible for the structural integrity of the virus are known for their conserved sequence compared to the S protein. These structural proteins are essential for performing the overall functions in the SARS-CoV-2 virus reproductive cycle [13]. The life cycle and the entire pathophysiological mechanism of SARS-CoV-2 virus are depicted in Figure 4 [20]. The surface spike protein supports the formation of the envelope, further pathogenesis, and the budding process as it helps in the encasing of the RNA and is also found to help in protein assembly [21,22].
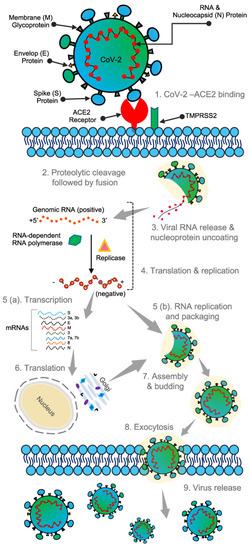
Figure 4. Structure and binding of SARS-CoV-2 virus to human cells. Reproduced with permission from [20], Creative Commons.
In the current pandemic, biomarkers are extremely important as they can enhance the production and approval of new and innovative drugs or vaccines. Biomarkers can be used to describe observable features of the disease and to determine the best mode of treatment based on the observed phenotypes and genotypes [23]. The most used targeted biomarkers in viral diagnostics are viral proteins observed on the viral envelope and viral genetic material such as RNA. For example, the major biomarker for SARS-CoV-2 detection is its genome [24,25]. The viral proteins encoded by the SARS-CoV-2 virus act as an alternative biomarker for viral detection theoretically but are not a practically viable option due to the complex nature of the proteins.
2.2. Symptoms of Long COVID
Symptoms of the SARS-CoV-2 virus are non-specific, and infected individuals can show no signs of infection (asymptomatic) or can show serious symptoms such as severe pneumonia, which can eventually lead to the infected patient’s death. A study of 41 patients diagnosed with COVID-19 had typical symptoms, including fever, cough, and fatigue [26]. In addition, some other symptoms, such as sputum, headache, hemoptysis, and diarrhea were also prevalent [26]. An important thing to note here is that all the patients taken under investigation exhibited pneumonia.
The symptoms of COVID-19 are similar to those of illnesses such as influenza. The three principal symptoms of COVID-19 are cough, fever, and shortness of breath. However, this list expanded as the virus mutated to include headache, fatigue, sore throat, and a loss of smell and taste. Studies indicate that patients older than 60 are at a higher risk than children, who are less likely to be infected. Elderly people have an increased risk of death compared to younger patients since the former are prone to several underlying diseases [27]. However, most of the information available about the prevalent symptoms of COVID-19 originates from studies that focus mainly on a limited section of the population who presented in a hospital setting [28].
2.3. SARS-CoV-2 RNA Detection
The SARS-CoV-2 virus consists of a single-stranded RNA that possesses a good genetic similarity to other members of the coronavirus family [29]. The main technique used for detection is RT-PCR, which amplifies the genetic material of the SARS-CoV-2 virus for testing. Genes targeted during RT-PCR include the spike protein, nucleocapsid, envelope genes, and RNA-dependent RNA polymerase [30]. There are two methods to isolate nucleic acids: the first is the lengthy extraction procedure, and the second is through direct capture from the sample [31].
After the isolation of the nucleic acids, PCR is carried out to amplify the target viral gene. Hybridization, such as using enzymatic assays, is one of the methods used for sensing viral nucleic acid. In the RT-PCR method, a nasopharyngeal swab is commonly utilized (although other routes are available) to take a sample from the patient under investigation, and RNA is extracted from the collected sample. This RNA is further reverse-transcribed into complementary DNA strands and amplified for detection using a fluorescence probe. Although RT-PCR methods are considered the standard for detecting the SARS-CoV-2 virus due to their high sensitivity, they have certain limitations. Hence, isothermal amplification or LAMP is used as an alternative. When optimized for detection, the RT-LAMP assay can be as sensitive as a PCR test.
2.4. SARS-CoV-2 Antigen Detection
Antigen-antibody interactions can be used to capture viral antigens by immobilizing the antibody responsible for capturing them on a sensor electrode. The spike protein, membrane protein, nucleocapsid protein, and envelope protein are used as targets for COVID-19 virus detection. The spike protein enters the host cell after its structure is evaluated, making it a promising candidate as a sensor target [33].
The most commonly used immune-based tests contain COVID-19-specific recombinant antigens that are initially immobilized on membranes made of nitrocellulose. Antibodies such as IgM and IgG are conjugated with colored latex beads and further immobilized onto different conjugate pads. The test sample initially encounters the nitrocellulose membrane, and the human antiviral antibodies will make conjugate complexes with colored antibodies.
This entry is adapted from the peer-reviewed paper 10.3390/bios12100890
This entry is offline, you can click here to edit this entry!
