Maize or corn (Zea mays L.), a plant species particularly generous in its production potential and in its wide diversity of uses, is fundamental for the development of a modern and efficient agriculture. Globally, maize ranks third in area and first in production, ahead of wheat and rice. Due to the large areas corn occupies, but especially to its yielding performance, maize is a major source of food for the world’s population. In addition to its nutritional importance for humans, corn is also basic food for animal feed and valuable raw material for industrial processing. The importance of corn for human health results from the fact that it is a food rich in nutrients, especially energy of 355 kcal per 100 g flour with 15% moisture, compared to 352 kcal for wheat flour, 348 kcal for rye flour, and 346 kcal for hulled barley. As food, maize also has some shortcomings, of which that the low amount of some essential amino acids was noted, such as lysine and tryptophan. Maize is also a valuable raw material for industry, extracting oil, starch, alcohol, glucose, and other products such as syrup, pectin, dextrin, plastics, lactic acid, acetic acid, acetone, dyes, and synthetic rubber from its grains. Paper, cardboard, and nitrocellulose can be made from corn stalks. Every part of the maize plant has economic worth, including the grain, leaves, stalk, tassel, and cob, which can be used to make a variety of food and non-food goods. Maize breeding research has traditionally concentrated on enhancing productive potential in newly created maize varieties because this criterion ensures a crop’s economic efficiency. Nowadays, genomics tools are essential for a precise, fast, and efficient breeding of crops especially in the context of climate challenges, but also may in the future represent a way to accelerate the processes of de novo domestication of the species.
- bioinformatics
- breeding by design
- genomic selection
- predictive breeding
- precision breeding
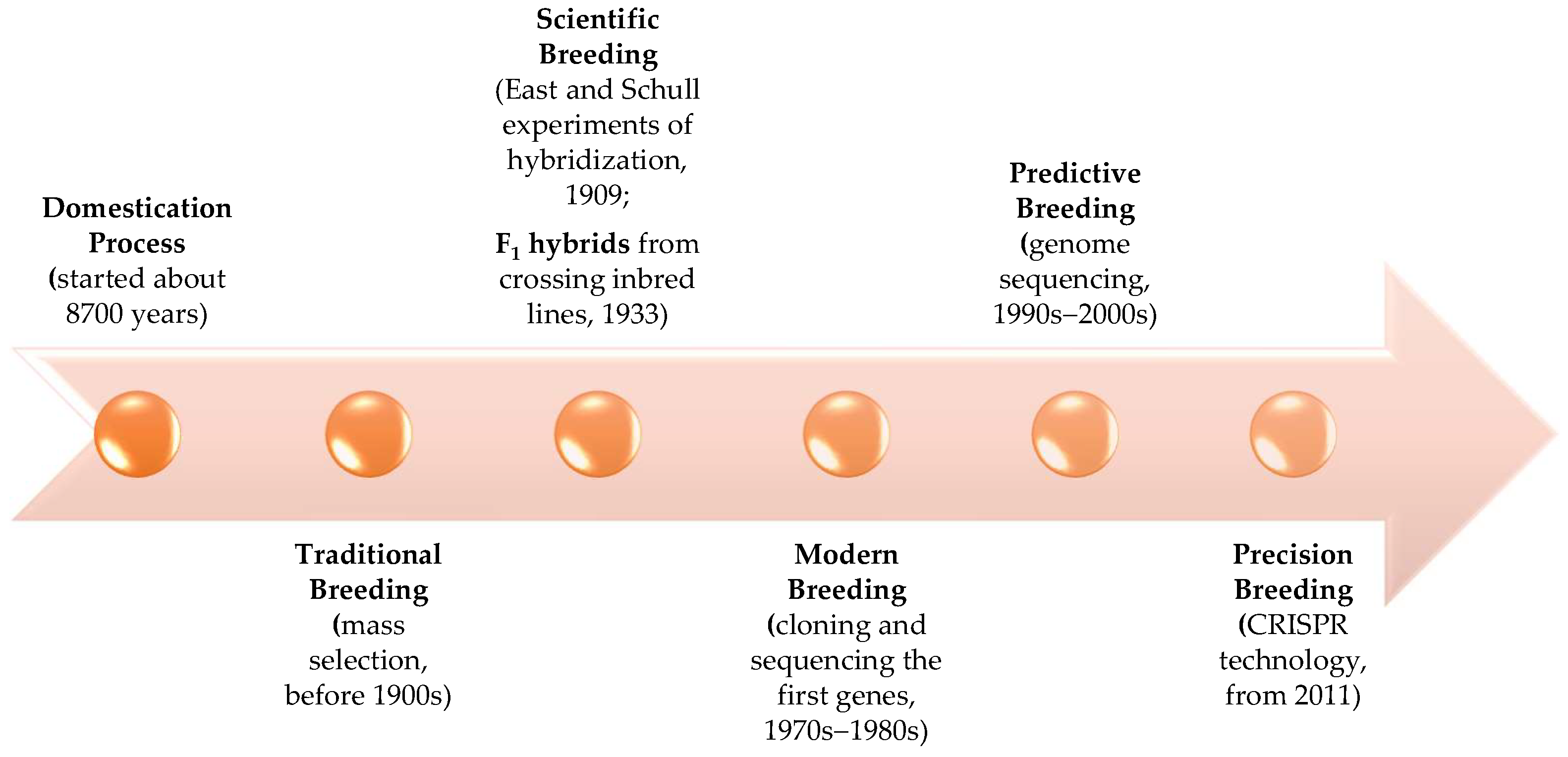
1. Evolution of Maize: From Domestication to Precise Breeding
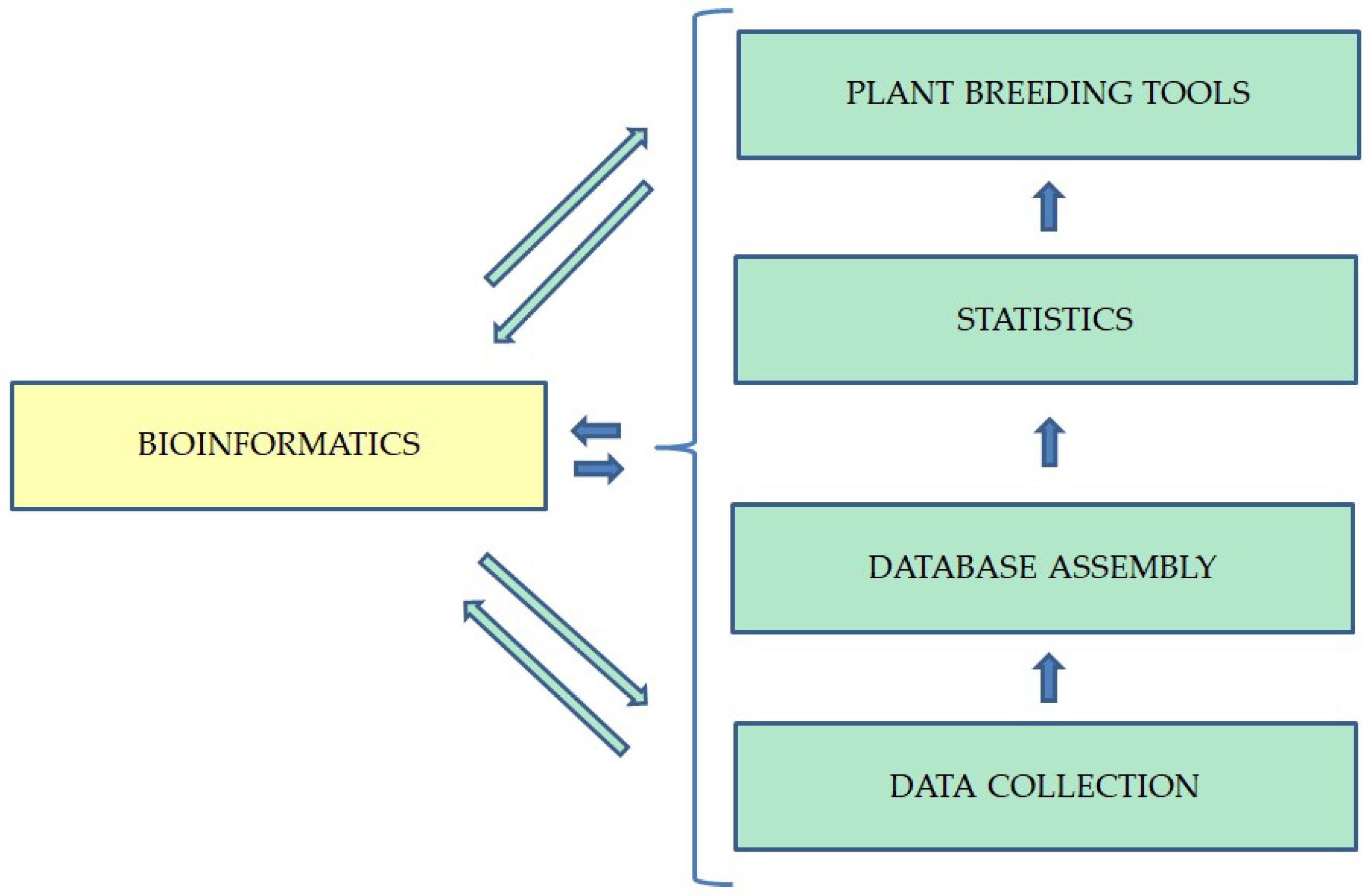
2. Bioinformatics: The Fundamental Tool Underlying Modern Methods of Breeding
3. Marker-Assisted Selection
4. Genomic Selection
5. Breeding by Design
6. Genome Sequencing
Genome sequencing (GSq) implies the determination of the order of DNA nucleotides in the genome of an organism [66]. Using this technique, scientists try to localize the genes and understand how a certain gene develops and influences the entire organism. In addition to the genes, GSq can help to a better understanding of the parts of genome outside the genes that control how genes are turned on or off [66]. Due to its complex genetics, maize is mostly used as a model plant for genetics analyses [67][68] and has become one of the first species considered for GSq [69]. It is estimated that maize genome is almost 80% of the human genome size and contains approximately 59,000 genes [70]. First sequencing attempts on maize were started during the early 2000s with The Sequencing the Maize Genome Project (STMG) [71][72] and The Consortium for Maize Genomics (CMG) [72][73]. A full-scale program was launched in 2005 by the National Science Foundation [72], to sequence the cultivar B73, one of the most used maize inbred lines for the production of the high-yielding commercial hybrids [74][75]. Three years later, the first draft sequence for maize genome was published [76]. According to a multi-genome investigation carried out in 2021, new variation in gene content, genome organization, and methylation was discovered in maize [77]. Genome prediction combined with crops modeling can lead to significant gains in maize breeding [32].
7. New Plant-Breeding Techniques
The start of the precision breeding was represented by the first actions of editing the plant genome [34]. Genome editing (GE) is the newest approach of modifying certain genes to improve the expression of interest traits. This technique can increase the genetic gain of the plants and also accelerate the breeding process at the same time [78]. GE modifications are precise, and the resulting changes are minor [79]. The GE experiments with high chances of success are the ones in which the target traits are determined by a single gene. The needed tools for GE are provided by the Novel Genomics Techniques (NGTs)/New Plant-Breeding Techniques (NBTs) [80]. GE instruments are based on DNA modification with the purpose of changing the traits of the plant [81][82]. Genome editing through NTBs implies using one the following methods: CRISPR/Cas, Zinc finger nuclease or TALENs [83][84][85][86][87][88], oligonucleotide-directed mutagenesis [89][90][91], epigenetic methods [92][93][94], cisgenesis [95][96][97], intragenesis [96][98][99], or grafting an unmodified plant with a transgenic one [100][101][102]. Using these types of tools combined with the classical breeding methods could remodel the maize breeding science [103] by speeding the entire process with shortening the generation time [104].
This entry is adapted from the peer-reviewed paper 10.3390/agronomy12102365
References
- Pérez-de-Castro, A.M.; Vilanova, S.; Cañizares, J.; Pascual, L.; Blanca, J.M.; Díez, M.J.; Prohens, J.; Picó, B. Application of Genomic Tools in Plant Breeding. Curr. Genom. 2012, 13, 179–195.
- Hake, S.; Ross-Ibarra, J. Genetic, Evolutionary and Plant Breeding Insights from the Domestication of Maize. eLife 2015, 4, e05861.
- Scott, M.P.; Emery, M. Maize: Overview. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2016; ISBN 978-0-08-100596-5.
- Haberer, G.; Young, S.; Bharti, A.K.; Gundlach, H.; Raymond, C.; Fuks, G.; Butler, E.; Wing, R.A.; Rounsley, S.; Birren, B.; et al. Structure and Architecture of the Maize Genome. Plant Physiol. 2005, 139, 1612–1624.
- Messing, J.; Bharti, A.K.; Karlowski, W.M.; Gundlach, H.; Kim, H.R.; Yu, Y.; Wei, F.; Fuks, G.; Soderlund, C.A.; Mayer, K.F.X.; et al. Sequence Composition and Genome Organization of Maize. Proc. Natl. Acad. Sci. USA 2004, 101, 14349–14354.
- Chandler, V.L.; Brendel, V. The Maize Genome Sequencing Project. Plant Physiol. 2002, 130, 1594–1597.
- Xu, Y.; Skinner, D.J.; Wu, H.; Palacios-Rojas, N.; Araus, J.L.; Yan, J.; Gao, S.; Warburton, M.L.; Crouch, J.H. Advances in Maize Genomics and Their Value for Enhancing Genetic Gains from Breeding. Int. J. Plant Genom. 2009, 2009, e957602.
- Palmer, L.E.; Rabinowicz, P.D.; O’Shaughnessy, A.L.; Balija, V.S.; Nascimento, L.U.; Dike, S.; de la Bastide, M.; Martienssen, R.A.; McCombie, W.R. Maize Genome Sequencing by Methylation Filtration. Science 2003, 302, 2115–2117.
- Mascher, M.; Gerlach, N.; Gahrtz, M.; Bucher, M.; Scholz, U.; Dresselhaus, T. Sequence and Ionomic Analysis of Divergent Strains of Maize Inbred Line B73 with an Altered Growth Phenotype. PLoS ONE 2014, 9, e96782.
- Stojaković, M.; Ivanović, M.; Bekavac, G.; Stojaković, Ž. Grain Yield of B73 x Mo17-Type Maize Hybrids from Different Periods of Breeding. Cereal Res. Commun. 2010, 38, 440–448.
- Stojaković, M.; Bekavac, G.; Vasić, N. B73 and Related Inbred Lines in Maize Breeding. Genetika 2005, 37, 245–252.
- Hufford, M.B.; Seetharam, A.S.; Woodhouse, M.R.; Chougule, K.M.; Ou, S.; Liu, J.; Ricci, W.A.; Guo, T.; Olson, A.; Qiu, Y.; et al. De Novo Assembly, Annotation, and Comparative Analysis of 26 Diverse Maize Genomes. Science 2021, 373, 105655–105662.
- Washburn, J.D.; Burch, M.B.; Franco, J.A.V. Predictive Breeding for Maize: Making Use of Molecular Phenotypes, Machine Learning, and Physiological Crop Models. Crop Sci. 2020, 60, 622–638.
- Kim, J.-I.; Kim, J.-Y. New Era of Precision Plant Breeding Using Genome Editing. Plant Biotechnol. Rep. 2019, 13, 419–421.
- Nepolean, T.; Kaul, J.; Mukri, G.; Mittal, S. Genomics-Enabled Next-Generation Breeding Approaches for Developing System-Specific Drought Tolerant Hybrids in Maize. Front. Plant Sci. 2018, 9, 361.
- Savadi, S.; Mangalassery, S.; Sandesh, M.S. Advances in Genomics and Genome Editing for Breeding next Generation of Fruit and Nut Crops. Genomics 2021, 113, 3718–3734.
- EU Regulation of New Plant Breeding Technologies and Their Possible Economic Implications for the EU and Beyond—Purnhagen—2021—Applied Economic Perspectives and Policy—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/full/10.1002/aepp.13084 (accessed on 30 March 2022).
- Eckerstorfer, M.F.; Engelhard, M.; Heissenberger, A.; Simon, S.; Teichmann, H. Plants Developed by New Genetic Modification Techniques—Comparison of Existing Regulatory Frameworks in the EU and Non-EU Countries. Front. Bioeng. Biotechnol. 2019, 7, 26.
- Enfissi, E.M.A.; Drapal, M.; Perez-Fons, L.; Nogueira, M.; Berry, H.M.; Almeida, J.; Fraser, P.D. New Plant Breeding Techniques and Their Regulatory Implications: An Opportunity to Advance Metabolomics Approaches. J. Plant Physiol. 2021, 258–259, 153378.
- Aglawe, S.B.; Barbadikar, K.M.; Mangrauthia, S.K.; Madhav, M.S. New Breeding Technique “Genome Editing” for Crop Improvement: Applications, Potentials and Challenges. 3 Biotech 2018, 8, 336.
- El-Mounadi, K.; Morales-Floriano, M.L.; Garcia-Ruiz, H. Principles, Applications, and Biosafety of Plant Genome Editing Using CRISPR-Cas9. Front. Plant Sci. 2020, 11, 56.
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for Crop Improvement: An Update Review. Front. Plant Sci. 2018, 9, 985.
- Petolino, J.F.; Kumar, S. Transgenic Trait Deployment Using Designed Nucleases. Plant Biotechnol. J. 2016, 14, 503–509.
- Smith, V.; Wesseler, J.H.H.; Zilberman, D. New Plant Breeding Technologies: An Assessment of the Political Economy of the Regulatory Environment and Implications for Sustainability. Sustainability 2021, 13, 3687.
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision Genome Editing in Plants: State-of-the-Art in CRISPR/Cas9-Based Genome Engineering. BMC Plant Biol. 2020, 20, 234.
- Lusser, M.; Parisi, C.; Plan, D.; Rodríguez-Cerezo, E. Deployment of New Biotechnologies in Plant Breeding. Nat. Biotechnol. 2012, 30, 231–239.
- Modrzejewski, D.; Hartung, F.; Sprink, T.; Krause, D.; Kohl, C.; Schiemann, J.; Wilhelm, R. What Is the Available Evidence for the Application of Genome Editing as a New Tool for Plant Trait Modification and the Potential Occurrence of Associated Off-Target Effects: A Systematic Map Protocol. Environ. Evid. 2018, 7, 18.
- Sauer, N.J.; Narváez-Vásquez, J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Woodward, M.J.; Mihiret, Y.A.; Lincoln, T.A.; Segami, R.E.; Sanders, S.L.; et al. Oligonucleotide-Mediated Genome Editing Provides Precision and Function to Engineered Nucleases and Antibiotics in Plants. Plant Physiol. 2016, 170, 1917–1928.
- Dalakouras, A.; Vlachostergios, D. Epigenetic Approaches to Crop Breeding: Current Status and Perspectives. J. Exp. Bot. 2021, 72, 5356–5371.
- Matzke, A.J.; Matzke, M.A. Position Effects and Epigenetic Silencing of Plant Transgenes. Curr. Opin. Plant Biol. 1998, 1, 142–148.
- Songstad, D.D.; Petolino, J.F.; Voytas, D.F.; Reichert, N.A. Genome Editing of Plants. Crit. Rev. Plant Sci. 2017, 36, 1–23.
- Cisgenesis and Genome Editing: Combining Concepts and Efforts for a Smarter Use of Genetic Resources in Crop Breeding—Cardi—2016—Plant Breeding—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/full/10.1111/pbr.12345 (accessed on 30 March 2022).
- Espinoza, C.; Schlechter, R.; Herrera, D.; Torres, E.; Serrano, A.; Medina, C.; Arce-Johnson, P. Cisgenesis and Intragenesis: New Tools for Improving Crops. Biol. Res. 2013, 46, 323–331.
- Hou, H.; Atlihan, N.; Lu, Z.-X. New Biotechnology Enhances the Application of Cisgenesis in Plant Breeding. Front. Plant Sci. 2014, 5, 389.
- Holme, I.B.; Wendt, T.; Holm, P.B. Intragenesis and Cisgenesis as Alternatives to Transgenic Crop Development. Plant Biotechnol. J. 2013, 11, 395–407.
- Súnico, V.; Higuera, J.J.; Molina-Hidalgo, F.J.; Blanco-Portales, R.; Moyano, E.; Rodríguez-Franco, A.; Muñoz-Blanco, J.; Caballero, J.L. The Intragenesis and Synthetic Biology Approach towards Accelerating Genetic Gains on Strawberry: Development of New Tools to Improve Fruit Quality and Resistance to Pathogens. Plants 2022, 11, 57.
- Song, G.; Walworth, A.E.; Loescher, W.H. Grafting of Genetically Engineered Plants. J. Am. Soc. Hortic. Sci. 2015, 140, 203–213.
- Kodama, H.; Miyahara, T.; Oguchi, T.; Tsujimoto, T.; Ozeki, Y.; Ogawa, T.; Yamaguchi, Y.; Ohta, D. Effect of Transgenic Rootstock Grafting on the Omics Profiles in Tomato. Food Saf. 2021, 9, 32–47.
- Tsaballa, A.; Xanthopoulou, A.; Madesis, P.; Tsaftaris, A.; Nianiou-Obeidat, I. Vegetable Grafting from a Molecular Point of View: The Involvement of Epigenetics in Rootstock-Scion Interactions. Front. Plant Sci. 2021, 11, 621999.
- Gedil, M.; Menkir, A. An Integrated Molecular and Conventional Breeding Scheme for Enhancing Genetic Gain in Maize in Africa. Front. Plant Sci. 2019, 10, 1430.
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed Breeding Is a Powerful Tool to Accelerate Crop Research and Breeding. Nat. Plants 2018, 4, 23–29.
- Genomic Selection for Crop Improvement—Heffner—2009—Crop Science—Wiley Online Library. Available online: https://acsess.onlinelibrary.wiley.com/doi/abs/10.2135/cropsci2008.08.0512 (accessed on 30 March 2022).
- Poland, J.A.; Rife, T.W. Genotyping-by-Sequencing for Plant Breeding and Genetics. Plant Genome 2012, 5, 92–102.
- Heslot, N.; Yang, H.-P.; Sorrells, M.E.; Jannink, J.-L. Genomic Selection in Plant Breeding: A Comparison of Models. Crop Sci. 2012, 52, 146–160.
- R2D2 Consortium; Fugeray-Scarbel, A.; Bastien, C.; Dupont-Nivet, M.; Lemarié, S. Why and How to Switch to Genomic Selection: Lessons from Plant and Animal Breeding Experience. Front. Genet. 2021, 12, 629737.
- Atanda, S.A.; Olsen, M.; Burgueño, J.; Crossa, J.; Dzidzienyo, D.; Beyene, Y.; Gowda, M.; Dreher, K.; Zhang, X.; Prasanna, B.M.; et al. Maximizing Efficiency of Genomic Selection in CIMMYT’s Tropical Maize Breeding Program. Theor. Appl. Genet. 2021, 134, 279–294.
- Robertsen, C.D.; Hjortshøj, R.L.; Janss, L.L. Genomic Selection in Cereal Breeding. Agronomy 2019, 9, 95.
- Pérez-Rodríguez, P.; Gianola, D.; González-Camacho, J.M.; Crossa, J.; Manès, Y.; Dreisigacker, S. Comparison Between Linear and Non-Parametric Regression Models for Genome-Enabled Prediction in Wheat. Genetics 2012, 2, 1595–1605.
- Windhausen, V.S.; Atlin, G.N.; Hickey, J.M.; Crossa, J.; Jannink, J.-L.; Sorrells, M.E.; Raman, B.; Cairns, J.E.; Tarekegne, A.; Semagn, K.; et al. Effectiveness of Genomic Prediction of Maize Hybrid Performance in Different Breeding Populations and Environments. Genetics 2012, 2, 1427–1436.
- Technow, F.; Bürger, A.; Melchinger, A.E. Genomic Prediction of Northern Corn Leaf Blight Resistance in Maize with Combined or Separated Training Sets for Heterotic Groups. Genetics 2013, 3, 197–203.
- Massman, J.M.; Jung, H.-J.G.; Bernardo, R. Genomewide Selection versus Marker-Assisted Recurrent Selection to Improve Grain Yield and Stover-Quality Traits for Cellulosic Ethanol in Maize. Crop Sci. 2013, 53, 58–66.
- Combs, E.; Bernardo, R. Genomewide Selection to Introgress Semidwarf Maize Germplasm into U.S. Corn Belt Inbreds. Crop Sci. 2013, 53, 1427–1436.
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975.
- Zhang, A.; Wang, H.; Beyene, Y.; Semagn, K.; Liu, Y.; Cao, S.; Cui, Z.; Ruan, Y.; Burgueño, J.; San Vicente, F.; et al. Effect of Trait Heritability, Training Population Size and Marker Density on Genomic Prediction Accuracy Estimation in 22 Bi-Parental Tropical Maize Populations. Front. Plant Sci. 2017, 8, 1916.
- Shepherd, R.K.; Meuwissen, T.H.; Woolliams, J.A. Genomic Selection and Complex Trait Prediction Using a Fast EM Algorithm Applied to Genome-Wide Markers. BMC Bioinform. 2010, 11, 529.
- Plant Breeding with Genomic Selection: Gain per Unit Time and Cost—Heffner—2010—Crop Science—Wiley Online Library. Available online: https://acsess.onlinelibrary.wiley.com/doi/abs/10.2135/cropsci2009.11.0662 (accessed on 30 March 2022).
- Jannink, J.-L.; Lorenz, A.J.; Iwata, H. Genomic Selection in Plant Breeding: From Theory to Practice. Brief. Funct. Genom. 2010, 9, 166–177.
- Beyene, Y.; Gowda, M.; Pérez-Rodríguez, P.; Olsen, M.; Robbins, K.R.; Burgueño, J.; Prasanna, B.M.; Crossa, J. Application of Genomic Selection at the Early Stage of Breeding Pipeline in Tropical Maize. Front. Plant Sci. 2021, 12, 685488.
- Hallauer, A.R.; Filho, J.B.M.; Carena, M.J. Breeding Plans. In Quantitative Genetics in Maize Breeding; Carena, M.J., Hallauer, A.R., Miranda Filho, J.B., Eds.; Handbook of Plant Breeding; Springer: New York, NY, USA, 2010; pp. 577–653. ISBN 978-1-4419-0766-0.
- Fritsche-Neto, R.; Akdemir, D.; Jannink, J.-L. Accuracy of Genomic Selection to Predict Maize Single-Crosses Obtained through Different Mating Designs. Theor. Appl. Genet. 2018, 131, 1153–1162.
- Rice, B.R.; Lipka, A.E. Diversifying Maize Genomic Selection Models. Mol. Breed. 2021, 41, 33.
- Xu, J. Breeding by Design for Future Rice: Genes and Genome Technologies. Crop J. 2021, 6, 491–496.
- Peleman, J.D.; van der Voort, J.R. Breeding by Design. Trends Plant Sci. 2003, 8, 330–334.
- Zhang, G. Target Chromosome-Segment Substitution: A Way to Breeding by Design in Rice. Crop J. 2021, 9, 658–668.
- Pérez-de-Castro, A.M.; Vilanova, S.; Cañizares, J.; Pascual, L.; Blanca, J.M.; Díez, M.J.; Prohens, J.; Picó, B. Application of Genomic Tools in Plant Breeding. Curr. Genom. 2012, 13, 179–195.
