The uncertain proportions of pandemic outbreaks have triggered the need of reliable and cost-effective protocols easily adaptable to the changing virulence of virus strains. In recent years, plasmonic biosensors are being increasingly applied for clinical diagnosis of viral and other infectious diseases. Typical plasmonic biosensing strategies rely on the versatility of SPR and LSPR as label-free detection systems capable of monitoring binding interactions in a short period of time. Nevertheless, the incorporation of technological advancements has precipitated the development of nanomaterial-based applications for improving the sensitivity and specificity of classical configurations. The unique optical properties of plasmonic nanostructures has been exploited in combination with SERS colorimetric, fluorescence or luminescence enhancement for viral diagnosis. Likewise, the development of plasmonic virus sensing approaches has also benefitted from the variety of virus biomarkers. Thus, a high number of virus plasmonic biosensors have prompted the advance of novel functionalization strategies to achieve the effective coverage of the biological receptor while ensuring the affinity and specificity towards the target viral nucleic acids, proteins or whole virus. The huge potential for single virus detection along with the effectiveness and simplicity of current plasmonic configurations will impact on the routine surveillance of virus in clinical settings during this decade.
- plasmonics
- virus detection
- nanomaterials
- SPR
- LSPR
- SERS
- colorimetric
- electrochemiluminescence
The global burden of coronavirus disease 2019 (COVID-19) to public health and global economy has stressed the need for rapid and simple diagnostic methods. From this perspective, plasmonic-based biosensing can manage the threat of infectious diseases by providing timely virus monitoring. In recent years, many plasmonics’ platforms have embraced the challenge of offering on-site strategies to complement traditional diagnostic methods relying on the polymerase chain reaction (PCR) and enzyme-linked immunosorbent assays (ELISA). This review compiled recent progress on the development of novel plasmonic sensing schemes for the effective control of virus-related diseases. A special focus was set on the utilization of plasmonic nanostructures in combination with other detection formats involving colorimetric, fluorescence, luminescence, or Raman scattering enhancement. The quantification of different viruses (e.g., hepatitis virus, influenza virus, norovirus, dengue virus, Ebola virus, Zika virus) with particular attention to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was reviewed from the perspective of the biomarker and the biological receptor immobilized on the sensor chip.
1. Introduction
Viral infections can be transmitted among human populations in a short period of time via droplets, aerosols, contact with abiotic surfaces, and even fecal matter [1][2][3]. The serious consequences of rapid airborne transmission are a cause of great concern since the global pandemic of coronavirus disease (COVID-19) has surpassed the number of cases and economic impact of recent virus disease outbreaks [e.g., H1N1/H5N1 flu, Ebola, MERS-CoV (Middle East Respiratory Syndrome Coronavirus), and SARS-CoV-1(Severe Acute Respiratory Syndrome Coronavirus 1)] [4]. The need for effective management of current and future epidemics has urged the development of rapid and sensitive screening tools. Therefore, viral diagnosis has become a primary need to control the spread of viral diseases [4][5][6][7][8][9].
Traditionally, viral diagnosis has relied upon culture-based methods and serological tests that detect either direct (intact viruses or their components: proteins or nucleic acids) or indirect (immunogens or antibodies against the viral antigens) virus infectious diseases [5].
More recently, the advances of molecular techniques in viral diagnosis have permitted that nucleic acid-based amplification methods such as the polymerase chain reaction (PCR) and the reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) have consolidated as the gold standard methods for the routine diagnosis of a wide range of viruses (human immunodeficiency virus (HIV), hepatitis B and C viruses, and cytomegalovirus (CMV)) [10][11][12][13]. In spite of the inherent advantages of PCR and RT-qPCR tests regarding sensitivity and selectivity, the utilization of time-consuming protocols (the report of results usually takes 1–3 days) and the high false-negative rate, limit their effectiveness to prevent the risk of new infections.
Consequently, the requirement for fast and cost-effective viral diagnostic methods has led to putting the focus on the development of real-time biosensing platforms. Among the variety of biosensor technologies that have arisen in the last decades for virus detection, plasmonic applications have gained significant attention, owing to their versatility, label-free monitoring, and low time of response [14]. The potential for multiplexing and system miniaturization are additional benefits for the point-of-care testing [15]. The combination of these features with the possibility of exploiting the physical and electronic properties of nanomaterials has allowed the design of ultrasensitive detection formats. In this way, novel plasmonic configurations taking advantage of colorimetric, surface-enhanced Raman scattering (SERS), and electrochemiluminescence sensing schemes seem to be ideally suited for monitoring either intact viruses or their components [16][17]. From this perspective, the fabrication of nanopatterned structures has emerged as a key factor for viral diagnosis using arrays of nanoplasmonic antennas [18]. The improvement in the spatial resolution of plasmonic substrates along with the enhancement of surface-to-volume ratios makes possible the detection of single virus particles with higher sensitivities [19].
On the other hand, the chemical activation of the surface plays a major role for improving the sensing efficiency of single virus particles [20][21]. The biofunctionalization of chemically activated substrates using aptamers, antibodies, antigens, or nucleic acids may enable the selective targeting of virus. Likewise, the design of effective surface coverages ensures the adequate orientation of biological receptors and, thereby, facilitate direct capture of intact virus from complex biological media.
Thus, nanoplasmonic biosensors provide a promising approach to attain ultra-low detection limits of viral particles, antigens, or nucleic acids from clinical specimens (i.e., blood, serum, saliva, etc.). The vast majority of virus sensing plasmonic applications are based on the well-known operation principles of surface plasmon resonance (SPR) biosensors. Nonetheless, the success in achieving optimal biosensing performance requires the design of novel biosensing strategies capable of maintaining the specificity and sensitivity of measurements while preserving the biocompatibility of the immobilized biological receptor.
Therefore, the aim of this review was to present recent advances in plasmonic biosensing schemes for the detection of virus on the basis of signal enhancement. This review specially focused on the utilization of sensing surfaces comprising nanomaterials-based approaches. Current progress in the development of plasmonic-based applications involving the combination with colorimetric, electrochemical, fluorescence, or fiber-optic detection principles is also discussed.
2. Biosensing Strategies
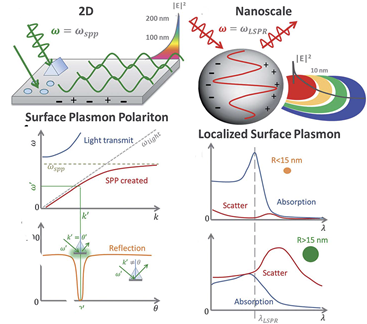
The fundamental principles of plasmonic biosensors rely on the propagation of surface plasmons along the interface of a thin, metal layer (commonly noble metal such as gold) and a dielectric (aqueous medium). A detailed description of the optical working principles was comprehensively reviewed in previous works and it is beyond the scope of this review [22][23][24]. In short, plasmonic biosensing takes advantage of the local refractive index changes of the transducer surface when monitoring molecular interactions between the target analyte and the immobilized biological receptor [25]. Binding events occurring in the surface can be monitored in two distinct forms: SPR and localized surface plasmon resonance (LSPR). Both SPR and LSPR depend on the refractive index of the surrounding media to induce spectral shifts. However, the dimension of the plasmonic nanomaterial determines the difference between SPR (metallic, thin layers) and LSPR [26], being below the wavelength of incident light in the latter (Figure 1).
Figure 1. Differences in volume, surface, and localized surface plasmon resonances of zero-dimensional metallic nanoparticles and two-dimensional thin, metallic surfaces. The surface plasmon polariton (SPP) can only be excited at certain wave vectors and decays evanescently from the surface. The momentum-matching condition leads to the SPP resonance and only exists at certain incident angles. In localized surface plasmon resonance, absorption dominates for small particles, less than ~15 nm, whereas the scattering cross-section dominates for big nanoparticles, greater than ~15 nm. Adapted with permission from Li et al. [26] Copyright © (2015) Royal Society of Chemistry (RSC)).
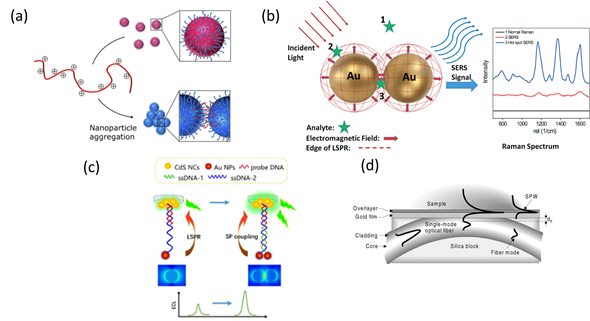
This special characteristic allows enhancing the spatial resolution of LSPR configurations by designing the geometry and composition of metallic nanostructures, laying the basis for colorimetric plasmonic biosensing [27][28]. The local electromagnetic field can enhance optical processes such as Raman scattering and fluorescence, leading to SERS and plasmon-enhanced electrochemiluminescence (ECL) sensing schemes [29][30][31]. The sensitivity achieved by both optical configurations is higher than that of SPR and LSPR, thus ensuring the detection of single virus particles. Alternatively, the propagation of electromagnetic radiation using optical fibers can also be applied to virus detection via miniaturized platforms with SPR and LSPR configurations [32][33] (Figure 2).
Figure 2. Schematic representation of plasmonic configurations: (a) Colorimetric assay showing dispersed (top) and aggregated (bottom) nanoparticles. Adapted with permission from Gormley et al. Copyright © (2014) American Chemical Society (ASC). (b) Surface-enhanced Raman spectroscopy (SERS) phenomenon for an organic analyte on gold nanoparticles (AuNPs). Adapted with permission from Wei et al. Copyright © (2015) RSC. (c) Schematic illustration of plasmonic-enhanced electrochemiluminescence (ECL) for DNA hybridization. Adapted with permission from Li et al. Copyright © (2016) RSC. (d) Surface Plasmon Resonance (SPR) sensing structure based on a side-polished, single-mode fiber. Adapted with permission from Slavík et al. Copyright © (2001) Elsevier.
Technological advancements in plasmonic biosensing including colorimetric and fluorescence enhancement as well as the utilization of nanomaterials and optical aperture nanostructures for achieving highly sensitive virus detection are described in this section.
2.1. Plasmonic Nanomaterials
Nanomaterials provide the signal amplification of plasmonic biosensing due to their interesting optical, magnetic, and electrical properties. The biocompatibility and easy chemical activation are also significant advantages. Likewise, the large surface area-to-volume ratio provides a higher loading of biological receptors [34][35][36]. Depending on their chemical composition, dimension, and physical properties, common types of plasmonic nanomaterials can vary from metallic nanoparticles and quantum dots to graphene nanostructures (i.e., carbon nanocomposites: Nanotubes, nanosheets, nanoflowers) [37][38][39]. Since the number of nanomaterial-based applications for viral sensing has increased significantly in the last decade, a comprehensive review of recent developments on plasmonic platforms is presented below and summarized in Table 1.
Table 1. Key analytical features of nanomaterials-based sensors classified according to the characteristics of target virus, detection format (namely immobilization strategy or biological receptor), and limit of detection.
|
Nanomaterial: Plasmonic Configuration |
Target Virus
|
Detection Format |
LOD (media/ biological sample) |
Reference |
|
Metal nanoparticles |
|
|
|
|
|
LSPR |
Hepatitis B surface antigen |
Hetero-assembled AuNPs sandwich-immunoassay |
100 fg mL−1 (human serum spiked samples) |
[40] |
|
Avian influenza virus (AIV H5N1) hemagglutinin protein |
Au spike-like nanoparticle onto indium-tin-oxide functionalized with DNA |
63.4 pg mL−1 (10% diluted chicken serum)1 |
||
|
Dengue NS1 antigen |
Thin silver film onto silicon substrate-Immunoassay/ Polyethersulfone membrane filter |
~0.06 μg mL−1 (Spiked whole blood samples) |
[42] |
|
|
Respiratory Syncytial Virus |
Antibody-functionalized gold, silver and copper nanoparticles/Intact virus |
2.4 plaque forming units (PFU) (eagle’s minimum essential medium) |
[43] |
|
|
SERS
|
Viral DNA (M13mp18single-stranded DNA) |
Nanoclusters of Au NP functionalized with Virus Like Particles |
0.25 ng μL−1 (bulk solution: water) |
[44] |
|
Hepatitis B surface antigen |
Composite gold nanorods and graphene sandwich-immunoassay |
0.05 pg mL−1 (9 patients diluted serum) |
||
|
Quantum dots |
|
|
|
|
|
LSPR-induced fluorescence
|
Influenza virus H1N1 antigens |
CdSeTeS QD functionalized with anti-neuraminidase antibody and AuNP conjugated to anti-hemagglutinin antibody-immunofluorescence assay |
0.03 pg mL−1 (deionized water) and 0.4 pg mL−1 (human serum) |
[46] |
|
Influenza virus H1N1 antigens |
CdZnSeS/ZnSeS QDs and gold nanoparticles- controlled distance and fluorescence quenching- immunofluorescence assay |
17.02 fg mL−1 (deionized water) and 65.1 fg mL−1 (10 % diluted human serum) |
[47] |
|
|
Norovirus |
Composites of Cysteine capped CdSeTeS QDs and AuNPs- fluorescence quenching- immunofluorescence assay |
12.1 × 10−15 g mL−1(10 % diluted human serum) |
[48] |
|
|
Zika RNA virus |
Nanohybrids of NPs bound to CdSeS alloyed QDs-hybridization |
2.4–7.6 copies mL−1 (assay buffer) |
||
|
Carbon-based |
|
|
|
|
|
SPR
|
Dengue virus (E-proteins, serotype 2) |
Composite of reduce graphene oxide and polyamidoamine (PAMAM) self-assembled to dithiobis (succinimidyl undecanoate) amine-activated layers- immunoassay antibody immobilization |
4.24 pg mL−1 (PBS solution)1 |
[50] |
|
Dengue virus (E-proteins, serotype 2) |
Cadmium sulfide quantum dots over graphene oxide |
53 pg mL−1 (PBS solution)1 |
[51] |
|
|
SERS magneto fluorometric
|
Influenza virus H1N1 hemagglutinin protein |
Binary AuNP-graphene hybrids and QD through antibody conjugated immunoassay a sandwich structure |
7.02 fg mL−1 (deionized water) 6.07 pg mL−1 (human serum) |
[52] |
|
Norovirus like particles |
Binary Metallic and magnetic nanoparticles decorated with graphene-immunoassay |
1.16 pg mL−1 (2% BSA solution) |
[53] |
|
|
Nanopatterning nanostructures |
|
|
|
|
|
Plasmonic fluorescence |
Ebola virus soluble glycoprotein |
3D plasmonic nanoantenna array Sandwich immunoassay format with fluorescent intensity enhancement |
220 fg mL−1 (diluted human plasma) |
[54] |
|
SPR |
Dengue virus like particles |
mPEG (polyethylene glycol)-SH-functionalized nanohole array. |
Not stated |
[55] |
|
EAR (extraordinary acoustic Raman) |
Virus particle of 25 nm (PhiX174) |
Optical trapping |
Not stated |
[56] |
1Values recalculated to express in mL; LOD: Limit of detection; AuNPs: Gold nanoparticles.
2.1.1. Metal Nanoparticles
Plasmonic nanomaterials can be classified regarding their chemical composition in organic (fullerenes, carbon nanotubes) and inorganic (metal, oxide-based nanomaterials, and quantum dots) materials. Among the latter ones, noble-metal nanoparticles have been extensively applied to plasmonic biosensing due to the strong absorption of light resulting from the oscillation of the free electrons. The size and shape of nanoparticles along with the interparticle distance determine the amplitude of oscillation and the position of the SPR band [57]. Likewise, variations of the chemical surrounding media due to the binding event between the biological receptor and the target analyte can also affect the oscillations. This implies that the physicochemical properties of the metallic nanoparticles can be tuned to transform LSPR spectra and produce even color variations, generating a signal response. Additionally, their high biocompatibility makes possible the functionalization of the surface with a broad range of biological receptors from antibodies to nucleic acids, glycoproteins, and aptamers, thus enabling the selective binding of a variety of analytes. Therefore, the utilization of gold and silver nanoparticles in LSPR, SERS, fluorescence enhancement, and colorimetric assays represents a popular approach for detecting small analytes.
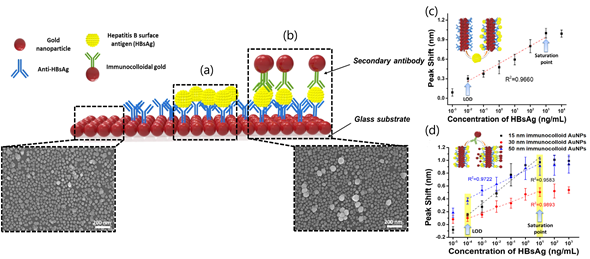
LSPR sensing is the most common sensing scheme that exploits noble-metal nanoparticles in analytical applications. LPSR biosensors employing gold nanoparticles (AuNPs) have been successfully applied to clinical diagnostics, environmental monitoring, and food safety. Particularly, a LSPR platform consisting of a hetero-assembled, AuNPs sandwich-immunoassay has been reported to detect hepatitis B surface antigen (HBsAg) at 100 fg mL−1. First, a glass substrate fabricated with AuNPs and conjugated with an anti-HBsAg antibody was prepared to detect target antigen HBsAg (Figure 3). After 10 min of inoculation, a second layer of AuNPs conjugated with anti-HBsAg antibody was formed to obtain the hetero-assembled, AuNPs sandwich-immunoassay chip format. The specificity of the immunoassay was tested against alpha fetoprotein (AFP), C-reactive protein (CRP), and prostate-specific antigen (PSA) while the validation was carried out in serum samples. Nevertheless, the suitability of the test for multiplexing analysis is suggested for further research.
Figure 3. Localized Surface Plasmon Resonance (LSPR) biosensing chip and analytical performance: (a) Gold nanoparticles arrayed on glass substrate; (b) modified hetero-assembled, AuNPs sandwich-immunoassay LSPR chip format; (c) detection of hepatitis B surface antigen (HBsAg) by single assay LSPR sensing chip format; and (d) modified hetero-assembled, AuNPs sandwich-immunoassay LSPR chip format using immunocolloid AuNPs. Absorbance spectra of both conjugated and unconjugated AuNPs were measured and compared in the wavelength range from 700 to 400 nm. Changes in the spectrum peak at different concentrations of HBsAg (1 pg mL−1 to 1 μg mL−1 HBsAg) were monitored using 15, 30, and 50 nm of immunocolloid AuNPs as signal enhancers. Adapted with permission from Kim et al. Copyright © (2018) Elsevier.
3. Plasmonic Advancements in COVID-19 Diagnosis
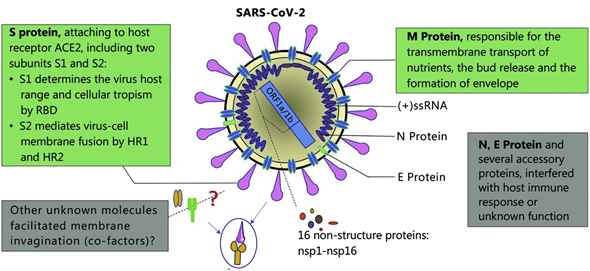
The SARS-CoV-2 is an enveloped, nonsegmented β-coronavirus that causes a new, severe, acute respiratory syndrome and coronavirus disease (COVID-19) . Since it is a positive-sense, single-stranded RNA virus (~30 kilobases in size and ~9860 amino acids), the genome of SARS-CoV-2 is the most important biomarker for diagnosis of COVID-19. SARS-CoV-2 structural and accessory proteins and even the whole virus SARS-CoV-2 could be used as antigens to monitor coronavirus disease (see Figure 4). Additionally, the detection of immunoglobulin M (IgM) and immunoglobulin G (IgG) responses after five days of the disease onset can also be utilized as indicators of COVID-19. As a result, reverse-transcription polymerase chain reaction (RT-PCR), gene sequencing, ELISA, and lateral flow immunoassay are the main diagnostic techniques at the moment [77] .
Figure 4. Schematic diagram of the Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) structure. Structural proteins, including spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein, and also several accessory proteins. Adapted with permission from Guo et al. [78] Copyright © (2020) BMC Springer Nature.
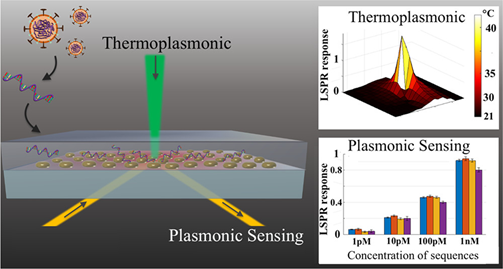
Nevertheless, the search for rapid and reliable point-of-care devices has triggered the development of plasmonic methods based on the latest technological advancements. For example, a novel approach combines the plasmonic photothermal (PPT) effect and LSPR sensing to detect DNA-selected sequences via hybridization to DNA receptors immobilized on two-dimensional gold nanoislands (AuNIs) (Figure 5). This dual-functional plasmonic biosensor takes advantage of the PPT heat generated on the AuNIs’ chip to increase the hybridization temperature and discriminate two similar gene sequences (RdRp genes) from SARS-CoV and SARS-CoV-2. A detection limit of 0.22 pM was obtained using a multigene mixture including the DNA sequences RdRp-COVID, open reading frame 1ab (ORF1ab)-COVID, and E genes from SARS-Cov-2. The thermoplasmonic enhancement also contributed to improve the stability of the assay by using two different angles of incidence to excite the plasmonic resonances of PPT and LSPR at two different wavelengths. Since the proposed method utilizes the same criteria for hybridization of nucleic acids, based on the decrease of the melting temperature than PCR-test, the potential to complement this technique is suggested.
Figure 5. Schematic representation of two-dimensional gold nanoislands functionalized with complementary DNA receptors, mapping the temperature distribution around the plasmonic photothermal (PPT) heat source and concentrations of various viral oligos measured using the dual-functional LSPR biosensors. An excitation laser with 532-nm peak wavelength and 40-mW maximum optical power was applied onto the gold nanoislands (AuNI) chip in the normal incident angle to improve the conversion efficiency of thermoplasmonic. Adapted with permission from Qiu et al. [75]Copyright © (2020) American Chemical Society.
Another innovative approach for COVID-19 diagnosis comprised the development of a colorimetric assay based on gold nanoparticles (AuNPs) functionalized with thiol-modified antisense oligonucleotides (ASOs) specific for N-gene (nucleocapsid phosphoprotein) of SARS-CoV-2 [76]. The biosensing scheme comprised the change in its SPR absorbance spectra with a red shift of ~40 nm when thiol-modified AuNPs agglomerated selectively in the presence of their target RNA sequence. This application also demonstrated that the addition of endonuclease Ribonuclease (RNAse H) leads to a visually detectable colorimetric change due to the agglomeration among the AuNPs, resulting from the cleaving of the RNA strand from the composite hybrid of RNA and Au-ASO composite. The assay selectivity was measured in the presence of MERS-CoV viral RNA, showing a limit of detection of 0.18 ng μL-1 of RNA with SARS-CoV-2 viral load. The main advantage of the proposed method was the possibility of being applied to target other regions of the viral genomic material, such as S-gene (surface glycoprotein), E-gene (envelope protein), and M-gene (membrane glycoprotein) without using sophisticated instrumental techniques.
This entry is adapted from the peer-reviewed paper 10.3390/s20174745
References
- Ong, S.W.X.; Tan, Y.K.; Chia, P.Y.; Lee, T.H.; Ng, O.T.; Wong, M.S.Y.; Marimuthu, K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA 2020, 323, 1610, doi:10.1001/jama.2020.3227.
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567, doi:10.1056/NEJMc2004973.
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844, doi:10.1001/jama.2020.3786.
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349, doi:10.1016/j.bios.2020.112349.
- Kumar, P. Monitoring Intact Viruses Using Aptamers. Biosensors 2016, 6, 40, doi:10.3390/bios6030040.
- Chen, H.; Liu, K.; Li, Z.; Wang, P. Point of care testing for infectious diseases. Clin. Chim. Acta 2019, 493, 138–147, doi:10.1016/j.cca.2019.03.008.
- Yanik, A.A.; Huang, M.; Kamohara, O.; Artar, A.; Geisbert, T.W.; Connor, J.H.; Altug, H. An Optofluidic Nanoplasmonic Biosensor for Direct Detection of Live Viruses from Biological Media. Nano Lett. 2010, 10, 4962–4969, doi:10.1021/nl103025u.
- Das, C.M.; Guo, Y.; Kang, L.; Ho, H.; Yong, K. Investigation of Plasmonic Detection of Human Respiratory Virus. Adv. Theory Simul. 2020, 3, 2000074, doi:10.1002/adts.202000074.
- Li, Z.; Leustean, L.; Inci, F.; Zheng, M.; Demirci, U.; Wang, S. Plasmonic-based platforms for diagnosis of infectious diseases at the point-of-care. Biotechnol. Adv. 2019, 37, 107440, doi:10.1016/j.biotechadv.2019.107440.
- Singh, P. Surface Plasmon Resonance: A Boon for Viral Diagnostics. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017; ISBN 978-0-12-809633-8.
- Mulder, J.; McKinney, N.; Christopherson, C.; Sninsky, J.; Greenfield, L.; Kwok, S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: Application to acute retroviral infection. J. Clin. Microbiol. 1994, 32, 292–300, doi:10.1128/JCM.32.2.292-300.1994.
- Berger, A.; Braner, J.; Doerr, H.W.; Weber, B. Quantification of Viral Load: Clinical Relevance for Human Immunodeficiency Virus, Hepatitis B Virus and Hepatitis C Virus Infection. Intervirology 1998, 41, 24–34, doi:10.1159/000024912.
- Boeckh, M.; Boivin, G. Quantitation of cytomegalovirus: Methodologic aspects and clinical applications. Clin. Microbiol. Rev. 1998, 11, 533–554.
- Lee, J.; Takemura, K.; Park, E. Plasmonic Nanomaterial-Based Optical Biosensing Platforms for Virus Detection. Sensors 2017, 17, 2332, doi:10.3390/s17102332.
- Qu, J.-H.; Dillen, A.; Saeys, W.; Lammertyn, J.; Spasic, D. Advancements in SPR biosensing technology: An overview of recent trends in smart layers design, multiplexing concepts, continuous monitoring and in vivo sensing. Anal. Chim. Acta 2020, 1104, 10–27, doi:10.1016/j.aca.2019.12.067.
- Kuttner, C. Plasmonics in Sensing: From Colorimetry to SERS Analytics. In Plasmonics; Gric, T., Ed.; IntechOpen: London, UK, 2018; ISBN 978-1-78984-434-4.
- Brolo, A.G. Plasmonics for future biosensors. Nat. Photon. 2012, 6, 709–713, doi:10.1038/nphoton.2012.266.
- Escobedo, C. On-chip nanohole array based sensing: A review. Lab Chip 2013, 13, 2445, doi:10.1039/c3lc50107h.
- Saha, K.; Agasti, S.S.; Kim, C.; Li, X.; Rotello, V.M. Gold Nanoparticles in Chemical and Biological Sensing. Chem. Rev. 2012, 112, 2739–2779, doi:10.1021/cr2001178.
- Oliverio, M.; Perotto, S.; Messina, G.C.; Lovato, L.; De Angelis, F. Chemical Functionalization of Plasmonic Surface Biosensors: A Tutorial Review on Issues, Strategies, and Costs. ACS Appl. Mater. Interfaces 2017, 9, 29394–29411, doi:10.1021/acsami.7b01583.
- De la Rica, R.; Stevens, M.M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat. Nanotechnol. 2012, 7, 821–824, doi:10.1038/nnano.2012.186.
- Homola, J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008, 108, 462–493, doi:10.1021/cr068107d.
- Masson, J.-F. Portable and field-deployed surface plasmon resonance and plasmonic sensors. Analyst 2020, 145, 3776–3800, doi:10.1039/D0AN00316F.
- Couture, M.; Zhao, S.S.; Masson, J.-F. Modern surface plasmon resonance for bioanalytics and biophysics. Phys. Chem. Chem. Phys. 2013, 15, 11190, doi:10.1039/c3cp50281c.
- Šípová, H.; Homola, J. Surface plasmon resonance sensing of nucleic acids: A review. Anal. Chim. Acta 2013, 773, 9–23, doi:10.1016/j.aca.2012.12.040.
- Li, M.; Cushing, S.K.; Wu, N. Plasmon-enhanced optical sensors: A review. Analyst 2015, 140, 386–406, doi:10.1039/C4AN01079E.
- Tang, L.; Li, J. Plasmon-Based Colorimetric Nanosensors for Ultrasensitive Molecular Diagnostics. ACS Sens. 2017, 2, 857–875, doi:10.1021/acssensors.7b00282.
- Gormley, A.J.; Chapman, R.; Stevens, M.M. Polymerization Amplified Detection for Nanoparticle-Based Biosensing. Nano Lett. 2014, 14, 6368–6373, doi:10.1021/nl502840h.
- Bantz, K.C.; Meyer, A.F.; Wittenberg, N.J.; Im, H.; Kurtuluş, Ö.; Lee, S.H.; Lindquist, N.C.; Oh, S.-H.; Haynes, C.L. Recent progress in SERS biosensing. Phys. Chem. Chem. Phys. 2011, 13, 11551, doi:10.1039/c0cp01841d.
- Wei, H.; Hossein Abtahi, S.M.; Vikesland, P.J. Plasmonic colorimetric and SERS sensors for environmental analysis. Environ. Sci. Nano 2015, 2, 120–135, doi:10.1039/C4EN00211C.
- Li, M.-X.; Zhao, W.; Qian, G.-S.; Feng, Q.-M.; Xu, J.-J.; Chen, H.-Y. Distance mediated electrochemiluminescence enhancement of CdS thin films induced by the plasmon coupling of gold nanoparticle dimers. Chem. Commun. 2016, 52, 14230–14233, doi:10.1039/C6CC08441A.
- Gauglitz, G. Critical assessment of relevant methods in the field of biosensors with direct optical detection based on fibers and waveguides using plasmonic, resonance, and interference effects. Anal. Bioanal. Chem. 2020, 412, 3317–3349, doi:10.1007/s00216-020-02581-0.
- Slavı́k, R.; Homola, J.; Čtyroký, J.; Brynda, E. Novel spectral fiber optic sensor based on surface plasmon resonance. Sens. Actuators B Chem. 2001, 74, 106–111, doi:10.1016/S0925-4005(00)00718-8.
- Doria, G.; Conde, J.; Veigas, B.; Giestas, L.; Almeida, C.; Assunção, M.; Rosa, J.; Baptista, P.V. Noble Metal Nanoparticles for Biosensing Applications. Sensors 2012, 12, 1657–1687, doi:10.3390/s120201657.
- Vikrant, K.; Bhardwaj, N.; Bhardwaj, S.K.; Kim, K.-H.; Deep, A. Nanomaterials as efficient platforms for sensing DNA. Biomaterials 2019, 214, 119215, doi:10.1016/j.biomaterials.2019.05.026.
- Choi, J.-H.; Lee, J.-H.; Son, J.; Choi, J.-W. Noble Metal-Assisted Surface Plasmon Resonance Immunosensors. Sensors 2020, 20, 1003, doi:10.3390/s20041003.
- Tran, V.T.; Zhou, H.; Kim, S.; Lee, J.; Kim, J.; Zou, F.; Kim, J.; Park, J.Y.; Lee, J. Self-assembled magnetoplasmonic nanochain for DNA sensing. Sens. Actuators B Chem. 2014, 203, 817–823, doi:10.1016/j.snb.2014.07.040.
- Adegoke, O.; Park, E.Y. Gold Nanoparticle-Quantum Dot Fluorescent Nanohybrid: Application for Localized Surface Plasmon Resonance-induced Molecular Beacon Ultrasensitive DNA Detection. Nanoscale Res. Lett. 2016, 11, 523, doi:10.1186/s11671-016-1748-3.
- Farzin, L.; Shamsipur, M.; Samandari, L.; Sheibani, S. HIV biosensors for early diagnosis of infection: The intertwine of nanotechnology with sensing strategies. Talanta 2020, 206, 120201, doi:10.1016/j.talanta.2019.120201.
- Kim, J.; Oh, S.Y.; Shukla, S.; Hong, S.B.; Heo, N.S.; Bajpai, V.K.; Chun, H.S.; Jo, C.-H.; Choi, B.G.; Huh, Y.S.; et al. Heteroassembled gold nanoparticles with sandwich-immunoassay LSPR chip format for rapid and sensitive detection of hepatitis B virus surface antigen (HBsAg). Biosens. Bioelectron. 2018, 107, 118–122, doi:10.1016/j.bios.2018.02.019.
- Lee, T.; Kim, G.H.; Kim, S.M.; Hong, K.; Kim, Y.; Park, C.; Sohn, H.; Min, J. Label-free localized surface plasmon resonance biosensor composed of multi-functional DNA 3 way junction on hollow Au spike-like nanoparticles (HAuSN) for avian influenza virus detection. Colloids Surfaces B Biointerfaces 2019, 182, 110341, doi:10.1016/j.colsurfb.2019.06.070.
- Austin Suthanthiraraj, P.P.; Sen, A.K. Localized surface plasmon resonance (LSPR) biosensor based on thermally annealed silver nanostructures with on-chip blood-plasma separation for the detection of dengue non-structural protein NS1 antigen. Biosens. Bioelectron. 2019, 132, 38–46, doi:10.1016/j.bios.2019.02.036.
- Valdez, J.; Bawage, S.; Gomez, I.; Singh, S.R. Facile and rapid detection of respiratory syncytial virus using metallic nanoparticles. J. Nanobiotechnol. 2016, 14, 13, doi:10.1186/s12951-016-0167-z.
- Lebedev, N.; Griva, I.; Dressick, W.J.; Phelps, J.; Johnson, J.E.; Meshcheriakova, Y.; Lomonossoff, G.P.; Soto, C.M. A virus-based nanoplasmonic structure as a surface-enhanced Raman biosensor. Biosens. Bioelectron. 2016, 77, 306–314, doi:10.1016/j.bios.2015.09.032.
- Liu, M.; Zheng, C.; Cui, M.; Zhang, X.; Yang, D.-P.; Wang, X.; Cui, D. Graphene oxide wrapped with gold nanorods as a tag in a SERS based immunoassay for the hepatitis B surface antigen. Microchim. Acta 2018, 185, 458, doi:10.1007/s00604-018-2989-x.
- Takemura, K.; Adegoke, O.; Takahashi, N.; Kato, T.; Li, T.-C.; Kitamoto, N.; Tanaka, T.; Suzuki, T.; Park, E.Y. Versatility of a localized surface plasmon resonance-based gold nanoparticle-alloyed quantum dot nanobiosensor for immunofluorescence detection of viruses. Biosens. Bioelectron. 2017, 89, 998–1005, doi:10.1016/j.bios.2016.10.045.
- Nasrin, F.; Chowdhury, A.D.; Takemura, K.; Kozaki, I.; Honda, H.; Adegoke, O.; Park, E.Y. Fluorometric virus detection platform using quantum dots-gold nanocomposites optimizing the linker length variation. Anal. Chim. Acta 2020, 1109, 148–157, doi:10.1016/j.aca.2020.02.039.
- Nasrin, F.; Chowdhury, A.D.; Takemura, K.; Lee, J.; Adegoke, O.; Deo, V.K.; Abe, F.; Suzuki, T.; Park, E.Y. Single-step detection of norovirus tuning localized surface plasmon resonance-induced optical signal between gold nanoparticles and quantum dots. Biosens. Bioelectron. 2018, 122, 16–24, doi:10.1016/j.bios.2018.09.024.
- Adegoke, O.; Morita, M.; Kato, T.; Ito, M.; Suzuki, T.; Park, E.Y. Localized surface plasmon resonance-mediated fluorescence signals in plasmonic nanoparticle-quantum dot hybrids for ultrasensitive Zika virus RNA detection via hairpin hybridization assays. Biosens. Bioelectron. 2017, 94, 513–522, doi:10.1016/j.bios.2017.03.046.
- Omar, N.A.S.; Fen, Y.W.; Abdullah, J.; Sadrolhosseini, A.R.; Mustapha Kamil, Y.; Fauzi, N.; ‘Illya, M.; Hashim, H.S.; Mahdi, M.A. Quantitative and Selective Surface Plasmon Resonance Response Based on a Reduced Graphene Oxide–Polyamidoamine Nanocomposite for Detection of Dengue Virus E-Proteins. Nanomaterials 2020, 10, 569, doi:10.3390/nano10030569.
- Omar, N.A.S.; Fen, Y.W.; Abdullah, J.; Zaid, M.H.M.; Daniyal, W.M.E.M.M.; Mahdi, M.A. Sensitive surface plasmon resonance performance of cadmium sulfide quantum dots-amine functionalized graphene oxide based thin film towards dengue virus E-protein. Opt. Laser Technol. 2019, 114, 204–208, doi:10.1016/j.optlastec.2019.01.038.
- Lee, J.; Takemura, K.; Park, E.Y. Plasmonic/magnetic graphene-based magnetofluoro-immunosensing platform for virus detection. Sens. Actuators B Chem. 2018, 276, 254–261, doi:10.1016/j.snb.2018.08.124.
- Lee, J.; Takemura, K.; Kato, C.N.; Suzuki, T.; Park, E.Y. Binary Nanoparticle Graphene Hybrid Structure-Based Highly Sensitive Biosensing Platform for Norovirus-Like Particle Detection. ACS Appl. Mater. Interfaces 2017, 9, 27298–27304, doi:10.1021/acsami.7b07012.
- Zang, F.; Su, Z.; Zhou, L.; Konduru, K.; Kaplan, G.; Chou, S.Y. Ultrasensitive Ebola Virus Antigen Sensing via 3D Nanoantenna Arrays. Adv. Mater. 2019, 1902331, doi:10.1002/adma.201902331.
- Jackman, J.A.; Linardy, E.; Yoo, D.; Seo, J.; Ng, W.B.; Klemme, D.J.; Wittenberg, N.J.; Oh, S.-H.; Cho, N.-J. Plasmonic Nanohole Sensor for Capturing Single Virus-Like Particles toward Virucidal Drug Evaluation. Small 2016, 12, 1159–1166, doi:10.1002/smll.201501914.
- Burkhartsmeyer, J.; Wang, Y.; Wong, K.S.; Gordon, R. Optical Trapping, Sizing, and Probing Acoustic Modes of a Small Virus. Appl. Sci. 2020, 10, 394, doi:10.3390/app10010394.
- Link, S.; El-Sayed, M.A. Size and Temperature Dependence of the Plasmon Absorption of Colloidal Gold Nanoparticles. J. Phys. Chem. B 1999, 103, 4212–4217, doi:10.1021/jp984796o.
- Lesiak, A.; Drzozga, K.; Cabaj, J.; Bański, M.; Malecha, K.; Podhorodecki, A. Optical Sensors Based on II-VI Quantum Dots. Nanomaterials 2019, 9, 192, doi:10.3390/nano9020192.
- Liu, S.; Zhao, N.; Cheng, Z.; Liu, H. Amino-functionalized green fluorescent carbon dots as surface energy transfer biosensors for hyaluronidase. Nanoscale 2015, 7, 6836–6842, doi:10.1039/C5NR00070J.
- Suvarnaphaet, P.; Pechprasarn, S. Graphene-Based Materials for Biosensors: A Review. Sensors 2017, 17, 2161, doi:10.3390/s17102161.
- Priyadarsini, S.; Mohanty, S.; Mukherjee, S.; Basu, S.; Mishra, M. Graphene and graphene oxide as nanomaterials for medicine and biology application. J. Nanostruct. Chem. 2018, 8, 123–137, doi:10.1007/s40097-018-0265-6.
- Escobedo, C.; Chou, Y.-W.; Rahman, M.; Duan, X.; Gordon, R.; Sinton, D.; Brolo, A.G.; Ferreira, J. Quantification of ovarian cancer markers with integrated microfluidic concentration gradient and imaging nanohole surface plasmon resonance. Analyst 2013, 138, 1450, doi:10.1039/c3an36616b.
- Gomez-Cruz, J.; Nair, S.; Manjarrez-Hernandez, A.; Gavilanes-Parra, S.; Ascanio, G.; Escobedo, C. Cost-effective flow-through nanohole array-based biosensing platform for the label-free detection of uropathogenic E. coli in real time. Biosens. Bioelectron. 2018, 106, 105–110, doi:10.1016/j.bios.2018.01.055.
- Jackman, J.A.; Ferhan, A.R.; Cho, N.-J. Surface-Based Nanoplasmonic Sensors for Biointerfacial Science Applications. BCSJ 2019, 92, 1404–1412, doi:10.1246/bcsj.20190112.
- Portela, A.; Calvo-Lozano, O.; Estevez, M.-C.; Medina Escuela, A.; Lechuga, L.M. Optical nanogap antennas as plasmonic biosensors for the detection of miRNA biomarkers. J. Mater. Chem. B 2020, 8, 4310–4317, doi:10.1039/D0TB00307G.
- Zhao, X.; Tsao, Y.-C.; Lee, F.-J.; Tsai, W.-H.; Wang, C.-H.; Chuang, T.-L.; Wu, M.-S.; Lin, C.-W. Optical fiber sensor based on surface plasmon resonance for rapid detection of avian influenza virus subtype H6: Initial studies. J. Virol. Methods 2016, 233, 15–22, doi:10.1016/j.jviromet.2016.03.007.
- Lin, H.-Y.; Huang, C.-H.; Lu, S.-H.; Kuo, I.-T.; Chau, L.-K. Direct detection of orchid viruses using nanorod-based fiber optic particle plasmon resonance immunosensor. Biosens. Bioelectron. 2014, 51, 371–378, doi:10.1016/j.bios.2013.08.009.
- Luo, B.; Xu, Y.; Wu, S.; Zhao, M.; Jiang, P.; Shi, S.; Zhang, Z.; Wang, Y.; Wang, L.; Liu, Y. A novel immunosensor based on excessively tilted fiber grating coated with gold nanospheres improves the detection limit of Newcastle disease virus. Biosens. Bioelectron. 2018, 100, 169–175, doi:10.1016/j.bios.2017.08.064.
- Lee, C.; Wang, P.; Gaston, M.A.; Weiss, A.A.; Zhang, P. Plasmonics-Based Detection of Virus Using Sialic Acid Functionalized Gold Nanoparticles. In Biosensors and Biodetection; Rasooly, A., Prickril, B., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1571, pp. 109–116, ISBN 978-1-4939-6846-6.
- Xu, S.; Ouyang, W.; Xie, P.; Lin, Y.; Qiu, B.; Lin, Z.; Chen, G.; Guo, L. Highly Uniform Gold Nanobipyramids for Ultrasensitive Colorimetric Detection of Influenza Virus. Anal. Chem. 2017, 89, 1617–1623, doi:10.1021/acs.analchem.6b03711.
- Khoris, I.M.; Chowdhury, A.D.; Li, T.-C.; Suzuki, T.; Park, E.Y. Advancement of capture immunoassay for real-time monitoring of hepatitis E virus-infected monkey. Anal. Chim. Acta 2020, 1110, 64–71, doi:10.1016/j.aca.2020.02.020.
- Miao, J.; Wang, J.; Guo, J.; Gao, H.; Han, K.; Jiang, C.; Miao, P. A plasmonic colorimetric strategy for visual miRNA detection based on hybridization chain reaction. Sci. Rep. 2016, 6, 32219, doi:10.1038/srep32219.
- Zhang, Q.; Liu, Y.; Nie, Y.; Ma, Q. Magnetic-plasmonic yolk-shell nanostructure-based plasmon-enhanced electrochemiluminescence sensor. Sens. Actuators B Chem. 2020, 319, 128245, doi:10.1016/j.snb.2020.128245.
- Liu, Y.; Nie, Y.; Wang, M.; Zhang, Q.; Ma, Q. Distance-dependent plasmon-enhanced electrochemiluminescence biosensor based on MoS2 nanosheets. Biosens. Bioelectron. 2020, 148, 111823, doi:10.1016/j.bios.2019.111823.
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277, doi:10.1021/acsnano.0c02439.
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627, doi:10.1021/acsnano.0c03822.
- Zhang, Z.; Wang, H.; Chen, Z.; Wang, X.; Choo, J.; Chen, L. Plasmonic colorimetric sensors based on etching and growth of noble metal nanoparticles: Strategies and applications. Biosens. Bioelectron. 2018, 114, 52–65, doi:10.1016/j.bios.2018.05.015.
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Military Med. Res. 2020, 7, 11, doi:10.1186/s40779-020-00240-0.