Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Methicillin-resistant Staphylococcus aureus (MRSA) belonging to several clonal complexes are widely distributed throughout the world. MRSA poses a serious threat to public health worldwide, due to the rapid spread and diversification of pandemic MRSA clones with increasing virulence and antimicrobial resistance.
- MRSA
- animals
- humans
- Staphylococcus aureus
1. Introduction
Staphylococcus aureus persistently or intermittently colonizes the nasal mucosa in approximately 30% of healthy adults [1]. Individuals naturally colonized by S. aureus have an increased risk of infection by this bacterium since its presence does not cause a detectable immune response in the host. In addition, these individuals are an important source of transmission of the microorganism to other people, which usually occurs through direct contact with the skin (Lee et al., 2018).
Antimicrobial resistance is a global public health problem that must be addressed by health authorities worldwide. Estimates from the European Union/European Economic Area show that each year more than 670,000 infections are due to antimicrobial-resistant bacteria with approximately 33,000 deaths [2].
The current methicillin-resistant Staphylococcus aureus (MRSA) situation in Europe has a prevalence of <5% in 9 European Union countries and >25% for 10 countries in the same region, namely Portugal [2]. In the hospital sector, resistant microorganisms deserve special attention. MRSA is one of the most frequent microorganisms that cause infections associated with the hospital environment [3]. In that same year, S. aureus were responsible for 18.5% of pneumonia episodes and 12% of bloodstream infections acquired in intensive care units in European hospitals. Of the S. aureus identified, 20% were MRSA [4]. In Portugal, 13.8% of pneumonia cases and 9.7% of bloodstream infections were caused by S. aureus strains [4]. The great variability in the rates of infections associated with the hospital environment in the various European countries is partly related to the different diagnostic methods. However, these data are of great importance as they alert to the need to implement measures to control and prevent the emergence and spread of resistant organisms.
Initially, MRSA infections were predominantly associated with the hospital environment, occurring mostly in hospitalized patients or patients who attended that environment. Therefore, during the 1960s−1970s, they were called HA-MRSA (Gajdács, 2019). However, in the late 1990s, MRSA infections began to appear in the community in people with no contact with the hospital environment and were therefore called CA-MRSA (Turner et al., 2019). These MRSA strains (HA-MRSA and CA-MRSA) belong to distinct genetic lineages and have some differences. HA-MRSA strains are generally more resistant to numerous drugs and have larger Staphylococcal Cassette Chromosome mec (SCCmec) types I, II and III. In contrast, CA-MRSA strains often have smaller SCCmec elements, usually types IV and V, and are not as resistant [5]. However, these strains appear to be more virulent due to the expression of virulence factors that increase their pathogenic potential [6], being one of the most frequent etiological agents of skin and soft tissue infections [7].
HA-MRSA are nosocomial and arise from wounds of infected patients, catheters and prolonged hospitalization, but also from the skin of healthy carriers [8]. In the late 1990s, MRSA infections began to appear in the community in people with no contact with the hospital environment [9]. The community-associated MRSA (CA-MRSA) has become a point of concern since these strains are associated with higher levels of virulence and disseminate fast, thus affecting seemingly healthy individuals [10]. In recent years, numerous MRSA strains have been isolated from different animal species, especially pigs, in many countries and were called livestock-associated MRSA (LA-MRSA) [11][12][13]. However, in most cases, MRSA colonized the animal host asymptomatically [14]. Most LA-MRSA belong to the genetic lineage clonal complex (CC) 398. Strains belonging to this same genetic lineage have been found in people who have direct contact with livestock [12][15][16]. In addition to this, vancomycin resistance has become a major concern within the scientific community. This adaptation arises from the acquisition of accessory components, in this case, the vanA gene [17], leading to the emergence of Vancomycin-resistant S. aureus (VRSA). This genetic adaptation is extremely important since there is a wide dependence on this antibiotic in the treatment of infections caused by MRSA. Vancomycin resistance in S. aureus tends to appear after prolonged or repeated periods of treatment with vancomycin, in a phenomenon called heteroresistance, where multiple mutations occur conferring different degrees of resistance to the antibiotic [18].
Currently, there has been an increase in the number of nosocomial infections by CA-MRSA; HA-MRSA clones have been responsible for infections in the community and LA-MRSA clones have been detected in healthy humans and human infections. Apparently, these clones have the ability to cross physical barriers, adapting easily. This change in the epidemiology of MRSA has raised serious concern as it becomes difficult to define the boundary between hospital–community-livestock transition.
2. Human-Associated Methicillin-resistant Staphylococcus Aureus
The control and prevention of infections by S. aureus strains can be carried out through rapid molecular typing that, simultaneously allows us to understand the transmission mechanism of this microorganism. MRSA typing can be performed through phenotypic or molecular characterization. In fact, the identification of endemic MRSA strains and strains responsible for disease outbreaks has become possible due to the improvement in molecular characterization techniques, which have allowed for a greater discrimination of MRSA clones [19]. Multilocus sequence typing (MLST) consists of the analysis of nucleotide sequences of internal fragments of seven housekeeping genes present in S. aureus. These genes are highly conserved since they encode enzymes necessary for the metabolism of the bacteria. The sequence of each locus is assigned an allele identification number based on its similarity to known alleles, and the combination of these seven alleles generates a sequence type (ST). When there are single nucleotide polymorphisms in fewer than three genes, the STs are considered closely related, and can be grouped under the same clonal complex (CC) [20]. The spa-typing technique is also widely used and it is based on the amplification and sequencing of the 24 bp X polymorphic zone of the spa gene, which encodes protein A, containing a variable number of repeats [21]. These repetitions are contrasted with those already known on an online server and, thus, the number associated with that strain of S. aureus can be determined.
MRSA strains known today resulted from processes of genetic recombination of pre-existing strains. Throughout this process of evolution, there was a selection of some advantageous characteristics that made these strains successful in a given geographic location [22][23]. Furthermore, the predominant lineage in that geographic location, after reaching a peak of dominance, tends to decline and later disappear, being replaced by a new lineage [24]. This clonal replacement process has been observed worldwide [25]. The acquisition of different types of Staphylococcal Cassette Chromosome mec (SCCmec) by methicillin-susceptible S. aureus (MSSA) strains of different genetic origins underlies the origin of MRSA pandemic clones such as those belonging to CC8: Archaic (ST250-I), Iberian (ST247-I), Brazilian (ST239-III) and USA300 (ST8-IV); and clones belonging to CC5, CC22, CC30 and CC398: New York/Japan (ST5-II), EMRSA-15 (ST22-IV), EMRSA-16 (ST36-II) and LA-MRSA (ST398-V), respectively [19][26].
MRSA infections in Europe in the early 1960s were limited to hospital outbreaks caused primarily by S. aureus with the phage 83A (later designated ST250). This clone was designated Archaic and was gradually replaced in the following decades by five other prevalent clones: CC5, CC8, CC22, CC30 and CC45 [6]. Of these, clones CC5 and CC8 are the most prevalent worldwide and comprise several different Sequence Types (ST’s) being widely distributed throughout the world (Table 1) [19].
| Clonal Complex (CC) | Sequence Type (ST) | Spa-Type | SCCmec Type | Clone | Geographic Distribution |
|---|---|---|---|---|---|
| CC1 | ST6 | t304 and variants | IVa | Middle East | The Middle East and Europe |
| CC5 | ST5 | t001, t002, t003, t010, t045, t053, t062, t105, t178, t179, t187, t214, t311, t319, t389, t443 | II | New York/Japan, USA100 | United States, Japan, Europe, Australia and South Korea |
| t001, t002, t003, t010, t045, t053, t062, t105, t178, t179, t187, t214, t311, t319, t389, t443 |
IV | Pediatric/USA800 | South America and Europe | ||
| CC8 | ST239 | t030, t037, t234, t387, t388 | III | Brazilian/Hungarian | Europe, South America, Asia and Africa |
| ST247 | t008, t051, t052, t054, t200 | I | Iberian/EMRSA-5 | Europe and the United States | |
| ST250 | t008, t009, t194 | I | Archaic | Worldwide | |
| CC22 | ST22 | t005, t022, t032, t223, t309, t310, t417, t420 | IV | EMRSA-15 | Europe, Australia and Canada |
| CC30 | ST36 | t018, t253, t418, t419 | II | EMRSA-16, USA200 | Europe, North America, and Australia |
| ST30 | t012, t019, t1143, t300 | IV | USA1100/South West Pacific | America, Australia and the Western Pacific | |
| CC45 | ST45 | t004, t015, t026, t031, t038, t050, t065, t204, t230, t390 | IV | Berlin, USA600 | Europe and the United States |
| ST80 | ST80 | t044, t203, t131, t1028, t1200 | IV | European | Europe, North Africa and the Middle East |
| ST93 | t3949 t202 t15361 t4699 t17089 t16949 t17272 | IV | Queensland | Australia |
Abbreviations. ST: sequence type; CC: clonal complex; SCCmec: Staphylococcal Cassette Chromosome mec.
Currently, Portugal has one of the highest nosocomial prevalence rates in Europe. Most HA-MRSA infections in Portugal are associated with EMRSA-15, the Iberian clone, and New York/Japan clones [29][30]. The Iberian clone was first described in Spain in 1989 and, since then, it has been reported in several countries, namely in Portugal. This, and the Portuguese clone (ST239-III variant) were the most prevalent clones of HA-MRSA from the mid-1980s to the beginning of the following decade [31][32]. The Pediatric clone was also mentioned for the first time in 1992, in a pediatric hospital in Portugal and was later found in Poland, the United States, Argentina and Colombia. Currently, the Clone New York/Japan is the most prevalent HA-MRSA clone in the country [19].
In addition to these, other clones such as the Brazilian (first described in 1992, in Brazil) have been identified worldwide. The New York/Japan clone, in turn, was identified as the dominant MRSA in hospitals in the metropolitan areas of New York, Pennsylvania, New Jersey and Connecticut in the United States, and also in a hospital in Tokyo, Japan [33]. However, in the last decade, there was a major shift in dominant MRSA clones from the New York/Japan to the USA300 clone [33]. In the United Kingdom, EMRSA-15 and EMRSA-16 were identified as endemic in hospitals in that region during the 1990s [34]. The same was true in several other European countries. Currently, EMRSA-15 is the dominant strain in UK hospitals, emphasizing the fact that CA-MRSA clones are widely disseminated in the hospital setting [9].
3. Animal-Associated Methicillin-resistant Staphylococcus Aureus
In addition to humans, MRSA also colonizes and infects animals. MRSA colonization and infection have been reported in a variety of animals, from domestic animals to farm animals. The indiscriminate use of antimicrobial agents in animal production and other agricultural activities has largely contributed to the distribution of MRSA among animals [35], thus, affecting more than 40% of pigs, 20% of cattle and up to 90% of turkey farms as demonstrated by a study carried out in Germany [36]. In Europe, LA-MRSA mostly belongs to the ST398 of the CC398, presenting itself as the largest reservoir of MRSA [37]. First recognized in 2005, LA-MRSA CC398 strains are the main type of MRSA reported in swine internationally and it is conceivable that this strain, which originated in swine, was later dispersed to other animals, such as cattle, horses and poultry [37][38][39][40][41]. While CC398 is the dominant lineage in livestock in Europe, other STs are predominant in other geographic locations. The dominant MRSA in livestock in Asia is CC9 [42]. Nevertheless, a wide range of genetic lineages have been reported in livestock such us CC1, CC5, CC9, CC45, CC97 and CC398 [43]. Figure 1 shows the distribution of MRSA CCs most commonly found in livestock and pets. MRSA strains often carried the mecA gene which confers resistance to methicillin but, the divergent mecA variant, mecC gene, has been increasingly reported in animals since it was first recognized in 2011 [44]. mecC-MRSA has been found in humans and animals, including pets, wild animals and livestock [11][45][46]. Most of the mecC-MRSA found among livestock belong to CC130 or CC425 [28]. Generally, MRSA clones of pets differ from those in livestock and meat production animals [47]. In cats and dogs, the most predominant MRSA lineage is ST22-IV (EMRSA-15) in Europe while in North America and Japan is ST5 [48][49]. As for horses, the most common lineages are ST8 and ST398 [41][50].
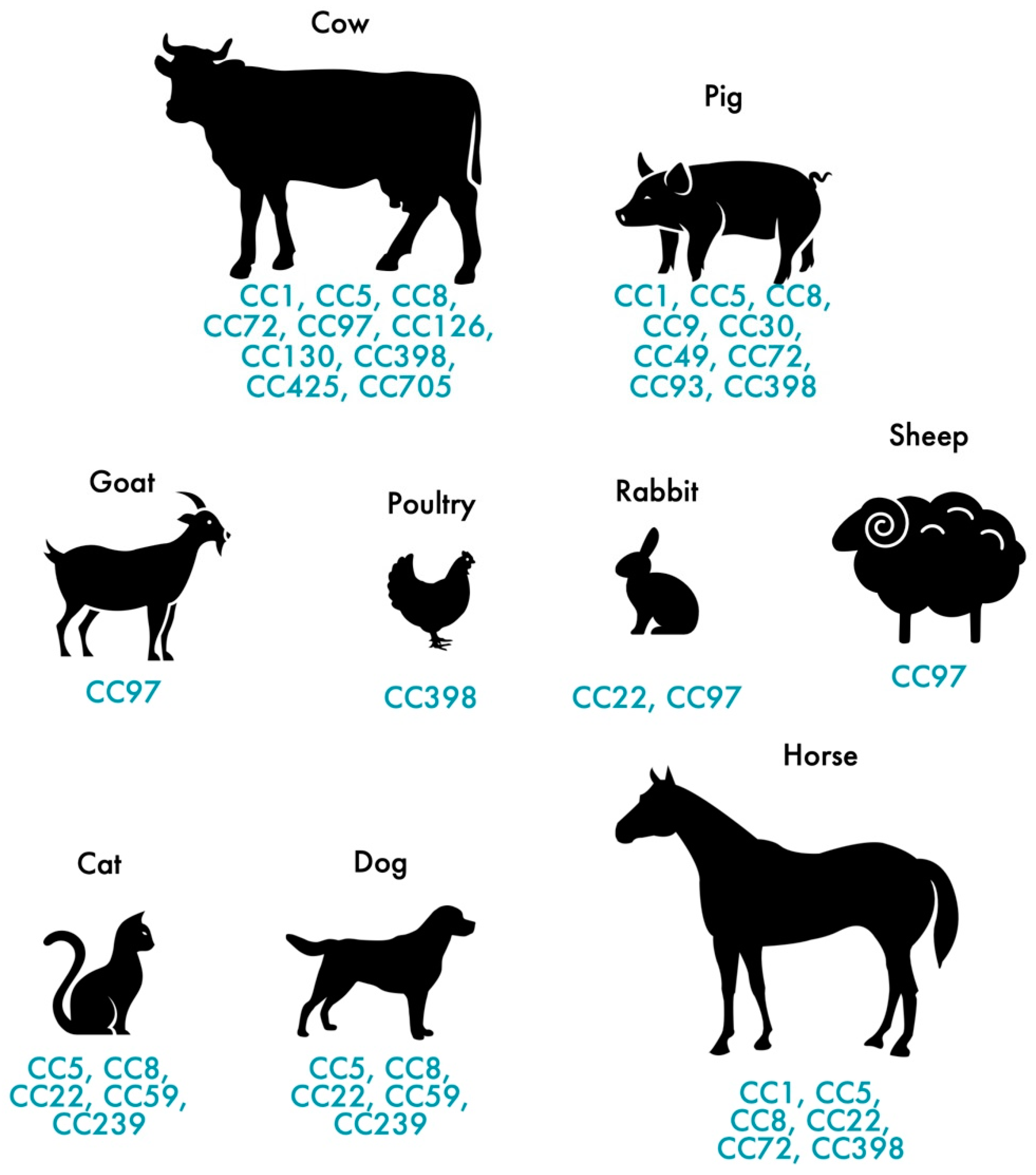
Figure 1. Distribution of the most frequent MRSA and MSSA clones in pets and livestock.
This entry is adapted from the peer-reviewed paper 10.3390/pathogens11101110
References
- González-García, S.; Hamdan-Partida, A.; Bustos-Hamdan, A.; Bustos-Martínez, J. Factors of Nasopharynx that Favor the Colonization and Persistence of Staphylococcus aureus. In Pharynx-Diagnosis and Treatment; IntechOpen: London, UK, 2021; ISBN 1789856094.
- European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2022–2020 Data; WHO Regional Office for Europe: Copenhagen, Swenden, 2022.
- Sankar, N.; Haja Abdul Nazeer, M.J.; Moinuddin, K. Spectrum of surgical site infections with special reference to methicillin resistant Staphylococcus aureus (MRSA). Int. J. Surg. 2020, 4, 306–309.
- European Centre for Disease Prevention and Control. Healthcare-associated infections acquired in intensive care units. In ECDC: Annual Epidemiological Report for 2017; ECDC: Stockholm, Sweden, 2019.
- Saïd-Salim, B.; Mathema, B.; Kreiswirth, B.N. Community-acquired methicillin-resistant Staphylococcus aureus: An emerging pathogen. Infect. Control Hosp. Epidemiol. 2003, 24, 451–455.
- Otto, M. MRSA virulence and spread. Cell. Microbiol. 2012, 14, 1513–1521.
- Liu, Y.; Xu, Z.; Yang, Z.; Sun, J.; Ma, L. Characterization of community-associated Staphylococcus aureus from skin and soft-tissue infections: A multicenter study in China. Emerg. Microbes Infect. 2016, 5, 1–11.
- Lee, A.S.; De Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Publ. Gr. 2018, 4, 1–23.
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218.
- Copin, R.; Sause, W.E.; Fulmer, Y.; Balasubramanian, D.; Dyzenhaus, S.; Ahmed, J.M.; Kumar, K.; Lees, J.; Stachel, A.; Fisher, J.C. Sequential evolution of virulence and resistance during clonal spread of community-acquired methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2019, 116, 1745–1754.
- Aklilu, E.; Chia, H.Y. First mecC and mecA Positive Livestock-Associated Methicillin Resistant Staphylococcus aureus (mecC MRSA/LA-MRSA) from Dairy Cattle in Malaysia. Microorganisms 2020, 8, 147.
- Sieber, R.N.; Larsen, A.R.; Urth, T.R.; Iversen, S.; Møller, C.H.; Skov, R.L.; Larsen, J.; Stegger, M. Genome investigations show host adaptation and transmission of LA-MRSA CC398 from pigs into Danish healthcare institutions. Sci. Rep. 2019, 9, 1–10.
- Bernier-Lachance, J.; Arsenault, J.; Usongo, V.; Parent, É.; Labrie, J.; Jacques, M.; Malouin, F.; Archambault, M. Prevalence and characteristics of Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) isolated from chicken meat in the province of Quebec, Canada. PLoS ONE 2020, 15, e0227183.
- Stone, M.J.; Swales, C.; Bond, S.E.; Muthayya, P.; Sarma, J.B. An outbreak of livestock-associated meticillin-resistant Staphylococcus aureus (LA-MRSA) clonal complex 398 in a regional burns centre. J. Hosp. Infect. 2022, 122, 1–8.
- Hansen, J.E.; Stegger, M.; Pedersen, K.; Sieber, R.N.; Larsen, J.; Larsen, G.; Lilje, B.; Chriél, M.; Andersen, P.S.; Larsen, A.R. Spread of LA-MRSA CC398 in Danish mink (Neovison vison) and mink farm workers. Vet. Microbiol. 2020, 245, 108705.
- Crespo-Piazuelo, D.; Lawlor, P.G. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir. Vet. J. 2021, 74, 21.
- Liu, W.-T.; Chen, E.-Z.; Yang, L.; Peng, C.; Wang, Q.; Xu, Z.; Chen, D.-Q. Emerging resistance mechanisms for 4 types of common anti-MRSA antibiotics in Staphylococcus aureus: A comprehensive review. Microb. Pathog. 2021, 156, 104915.
- Basco, M.D.S.; Kothari, A.; McKinzie, P.B.; Revollo, J.R.; Agnihothram, S.; Azevedo, M.P.; Saccente, M.; Hart, M.E. Reduced vancomycin susceptibility and increased macrophage survival in Staphylococcus aureus strains sequentially isolated from a bacteraemic patient during a short course of antibiotic therapy. J. Med. Microbiol. 2019, 68, 848–859.
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18.
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641.
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of Methicillin-Resistant Staphylococcus aureus in a University Hospital Setting by Using Novel Software for spa Repeat Determination and Database Management. J. Clin. Microbiol. 2003, 41, 5442–5448.
- Uhlemann, A.; McAdam, P.R.; Sullivan, S.B.; Knox, J.R.; Khiabanian, H.; Rabadan, R.; Davies, P.R.; Fitzgerald, J.R.; Lowy, F.D. Evolutionary Dynamics of Pandemic Methicillin-Sensitive Staphylococcus aureus ST398 and Its International Spread via Routes of Human Migration. MBio 2022, 8, e01375-16.
- Carrel, M.; Perencevich, E.N.; David, M.Z. USA300 Methicillin-Resistant Staphylococcus aureus, United States, 2000–2013. Emerg. Infect. Dis. 2015, 21, 1973–1980.
- Dai, Y.; Liu, J.; Guo, W.; Meng, H.; Huang, Q.; He, L.; Gao, Q.; Lv, H.; Liu, Y.; Wang, Y.; et al. Decreasing methicillin-resistant Staphylococcus aureus (MRSA) infections is attributable to the disappearance of predominant MRSA ST239 clones, Shanghai, 2008–2017. Emerg. Microbes Infect. 2019, 8, 471–478.
- Rolain, J.-M.; Abat, C.; Brouqui, P.; Raoult, D. Worldwide decrease in methicillin-resistant Staphylococcus aureus: Do we understand something? Clin. Microbiol. Infect. 2015, 21, 515–517.
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A Field Guide to Pandemic, Epidemic and Sporadic Clones of Methicillin-Resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936.
- Stefani, S.; Chung, D.R.; Lindsay, J.A.; Friedrich, A.W.; Kearns, A.M.; Westh, H.; MacKenzie, F.M. Meticillin-resistant Staphylococcus aureus (MRSA): Global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 2012, 39, 273–282.
- Bal, A.M.; Coombs, G.W.; Holden, M.T.G.; Lindsay, J.A.; Nimmo, G.R.; Tattevin, P.; Skov, R.L. Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: Blurring of the traditional definitions. J. Glob. Antimicrob. Resist. 2016, 6, 95–101.
- Silva, V.; Almeida, F.; Carvalho, J.A.; Castro, A.P.; Ferreira, E.; Manageiro, V.; Tejedor-Junco, M.T.; Caniça, M.; Igrejas, G.; Poeta, P. Emergence of community-acquired methicillin-resistant Staphylococcus aureus EMRSA-15 clone as the predominant cause of diabetic foot ulcer infections in Portugal. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 179–186.
- Silva, V.; Hermenegildo, S.; Ferreira, C.; Manaia, C.M.; Capita, R.; Alonso-Calleja, C.; Carvalho, I.; Pereira, J.E.; Maltez, L.; Capelo, J.L. Genetic Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Human Bloodstream Infections: Detection of MLSB Resistance. Antibiotics 2020, 9, 375.
- Oliveira, D.; Santos-Sanches, I.; Mato, R.; Tamayo, M.; Ribeiro, G.; Costa, D.; De Lencastre, H. Virtually all methicillin-resistant Staphylococcus aureus (MRSA) infections in the largest Portuguese teaching hospital are caused by two internationally spread multiresistant strains: The ‘Iberian’and the ‘Brazilian’clones of MRSA. Clin. Microbiol. Infect. 1998, 4, 373–384.
- De Sousa, M.A.; de Lencastre, H. Bridges from hospitals to the laboratory: Genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol. Med. Microbiol. 2004, 40, 101–111.
- Pardos de la Gandara, M.; Curry, M.; Berger, J.; Burstein, D.; Della-Latta, P.; Kopetz, V.; Quale, J.; Spitzer, E.; Tan, R.; Urban, C.; et al. MRSA Causing Infections in Hospitals in Greater Metropolitan New York: Major Shift in the Dominant Clonal Type between 1996 and 2014. PLoS ONE 2016, 11, e0156924.
- Moore, P.C.L.; Lindsay, J.A. Molecular characterisation of the dominant UK methicillin-resistant Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 2002, 51, 516–521.
- Van, T.T.H.; Yidana, Z.; Smooker, P.M.; Coloe, P.J. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J. Glob. Antimicrob. Resist. 2020, 20, 170–177.
- Köck, R.; Ballhausen, B.; Bischoff, M.; Cuny, C.; Eckmanns, T.; Fetsch, A.; Harmsen, D.; Goerge, T.; Oberheitmann, B.; Schwarz, S.; et al. The impact of zoonotic MRSA colonization and infection in Germany. Berl. Munch. Tierarztl. Wochenschr. 2014, 127, 384–398.
- Silva, V.; Vieira-Pinto, M.; Saraiva, C.; Manageiro, V.; Reis, L.; Ferreira, E.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Prevalence and Characteristics of Multidrug-Resistant Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from Quails (Coturnix Coturnix Japonica) Slaughtered for Human Consumption. Animals 2021, 11, 2038.
- Armand-Lefevre, L.; Ruimy, R.; Andremont, A. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 2005, 11, 711.
- Voss, A.; Loeffen, F.; Bakker, J.; Klaassen, C.; Wulf, M. Methicillin-resistant Staphylococcus aureus in Pig Farming. Emerg. Infect. Dis. 2005, 11, 1965–1966.
- Hansen, J.E.; Ronco, T.; Stegger, M.; Sieber, R.N.; Fertner, M.E.; Martin, H.L.; Farre, M.; Toft, N.; Larsen, A.R.; Pedersen, K. LA-MRSA CC398 in dairy cattle and veal calf farms indicates spillover from pig production. Front. Microbiol. 2019, 10, 2733.
- Schmitt, S.; Stephan, R.; Huebschke, E.; Schaefle, D.; Merz, A.; Johler, S. DNA microarray-based characterization and antimicrobial resistance phenotypes of clinical MRSA strains from animal hosts. J. Vet. Sci. 2020, 21, e54.
- Chen, C.-J.; Lauderdale, T.-L.Y.; Lu, C.-T.; Chuang, Y.-Y.; Yang, C.-C.; Wu, T.-S.; Lee, C.-Y.; Lu, M.-C.; Ko, W.-C.; Huang, Y.-C. Clinical and molecular features of MDR livestock-associated MRSA ST9 with staphylococcal cassette chromosome mec XII in humans. J. Antimicrob. Chemother. 2018, 73, 33–40.
- McCarthy, A.J.; Lindsay, J.A.; Loeffler, A. Are all meticillin-resistant Staphylococcus aureus (MRSA) equal in all hosts? Epidemiological and genetic comparison between animal and human MRSA. Vet. Dermatol. 2012, 23, 267-e54.
- Shore, A.C.; Deasy, E.C.; Slickers, P.; Brennan, G.; O’Connell, B.; Monecke, S.; Ehricht, R.; Coleman, D.C. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 3765–3773.
- Kerschner, H.; Harrison, E.M.; Hartl, R.; Holmes, M.A.; Apfalter, P. First report of mecC MRSA in human samples from Austria: Molecular characteristics and clinical data. New Microbes New Infect. 2015, 3, 4–9.
- Lindgren, A.-K.; Gustafsson, E.; Petersson, A.C.; Melander, E. Methicillin-resistant Staphylococcus aureus with mecC: A description of 45 human cases in southern Sweden. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 971–975.
- Haag, A.F.; Fitzgerald, J.R.; Penadés, J.R. Staphylococcus aureus in Animals. Microbiol. Spectr. 2019, 7, 7-3.
- Taniguchi, Y.; Koide, S.; Maeyama, Y.; Tamai, K.; Hayashi, W.; Tanaka, H.; Iimura, M.; Suzuki, M.; Nagano, Y.; Arakawa, Y.; et al. Predominance of methicillin-resistant Staphylococcus aureus SCCmec type II-CC5 and SCCmec type IV-CC1/CC8 among companion animal clinical isolates in Japan: Findings from phylogenetic comparison with human clinical isolates. J. Glob. Antimicrob. Resist. 2020, 20, 253–259.
- Matuszewska, M.; Murray, G.G.R.; Harrison, E.M.; Holmes, M.A.; Weinert, L.A. The Evolutionary Genomics of Host Specificity in Staphylococcus aureus. Trends Microbiol. 2020, 28, 465–477.
- Kaspar, U.; von Lützau, K.; Schlattmann, A.; Rösler, U.; Köck, R.; Becker, K. Zoonotic multidrug-resistant microorganisms among non-hospitalized horses from Germany. One Health 2019, 7, 100091.
This entry is offline, you can click here to edit this entry!
