Deep learning (DL) has become a prominent issue in the machine learning (ML) domain in the past few years. ML can be utilized to tackle issues in different sectors. Neuroscience is included in this list. It is well known that detecting malignancies and functioning regions in cognitive systems has been a huge challenge for scientists over the years. The standard approach of detecting the variation in blood oxygen levels can be applied for this purpose. However, completing all the processes can take too long on certain occasions. One benefit of DL approaches over typical ML methods is that the reliability of DL techniques grows with the phases of learning. The efficiency of DL methods tends to rise greatly as more information is provided to them, and they outperform conventional techniques. This is similar to the human brain, which learns more as new information becomes available on a daily basis.
- deep learning
- health informatics
- Alzheimer’s disease
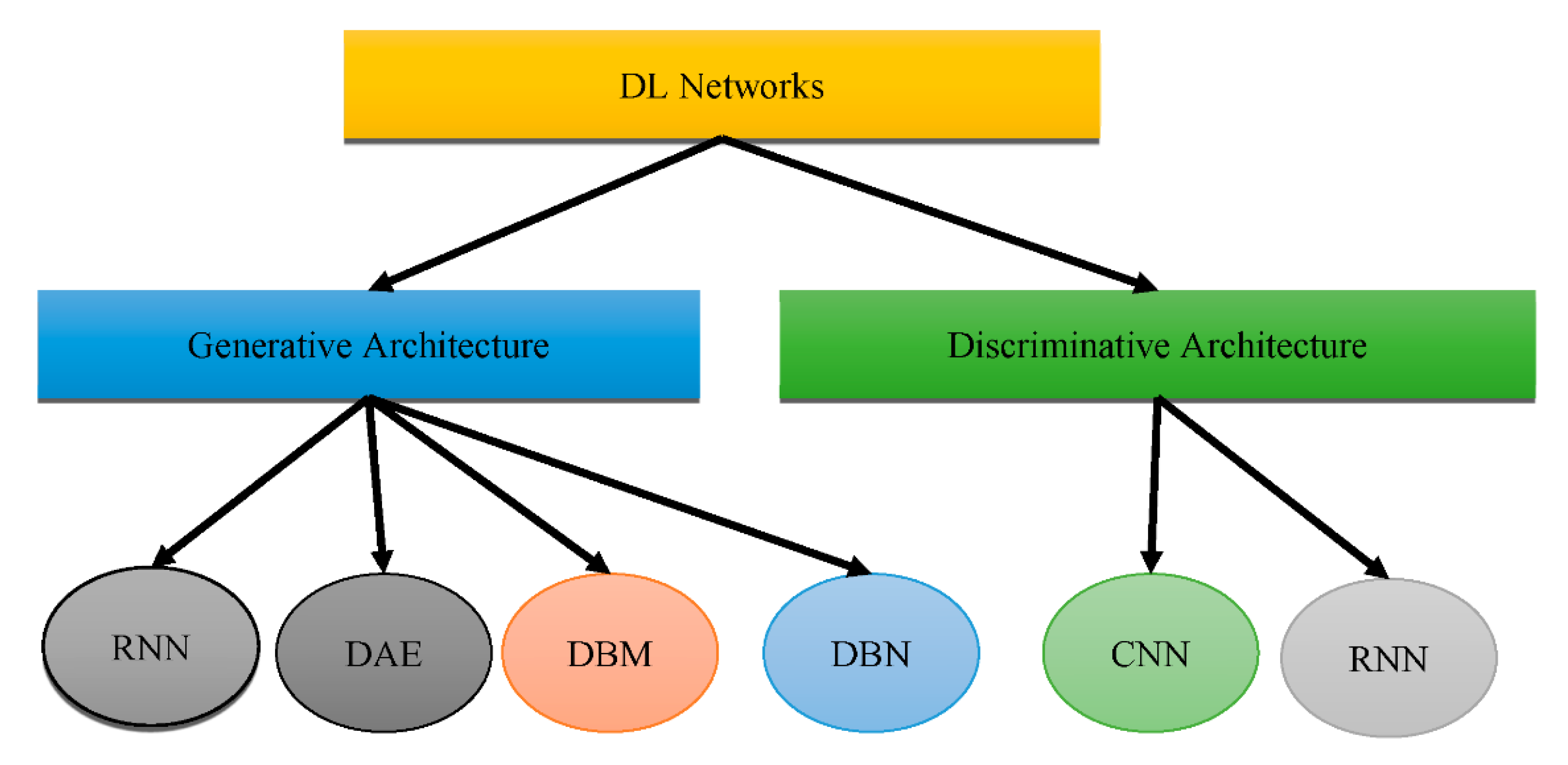
1. Transformation from Machine Learning (ML) to Deep Learning (DL) Approaches for the Effective Prediction of Alzheimer’s Disease (AD)
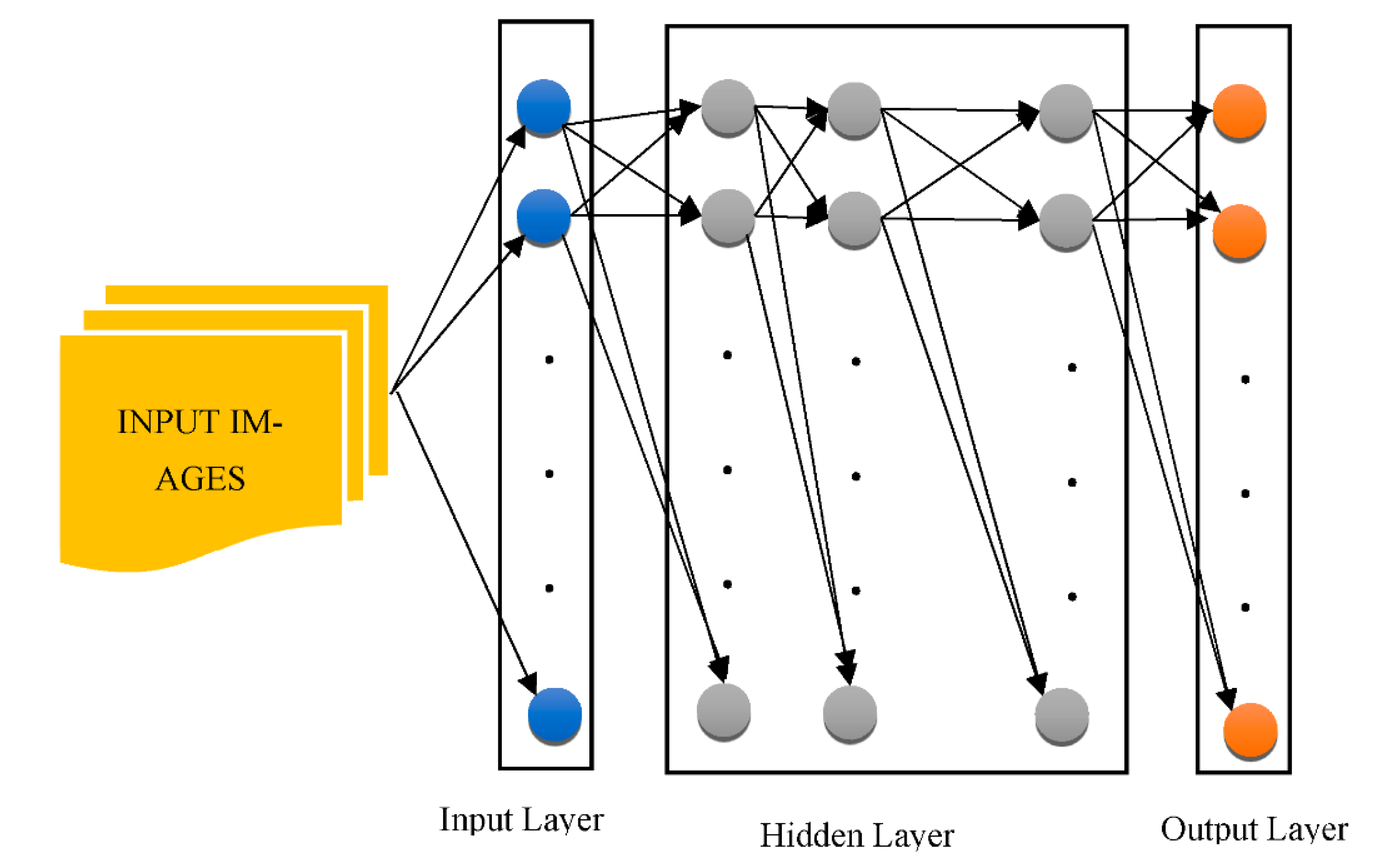
-
“Free Surfer” is an application for cerebral localization with cortex-associated information.
-
The “SPM5 (Statistical Parametric Mapping Tool)” is a device for the mapping of statistical parameters.
2. Diagnosis and Prognosis of AD Using DL Methods
This entry is adapted from the peer-reviewed paper 10.3390/healthcare10101842
References
- Al-Shoukry, S.; Rassem, T.H.; Makbol, N.M. Alzheimer’s Diseases Detection by Using Deep Learning Algorithms: A Mini-Review. IEEE Access 2020, 8, 77131–77141.
- Haller, S.; Nguyen, D.; Rodriguez, C.; Emch, J.; Gold, G.; Bartsch, A.; Lovblad, K.O.; Giannakopoulos, P. Individual prediction of cognitive decline in mild cognitive impairment using support vector machine-based analysis of diffusion tensor imaging data. J. Alzheimer’s Dis. 2010, 22, 315–327.
- Gamarra, M.; Mitre-Ortiz, A.; Escalante, H. Automatic cell image segmentation using genetic algorithms. In Proceedings of the 2019 XXII Symposium on Image, Signal Processing and Artificial Vision (STSIVA), Bucaramanga, Colombia, 24–26 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5.
- Fogel, I.; Sagi, D. Gabor filters as texture discriminator. Biol. Cybern. 1989, 61, 103–113.
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621.
- Deng, L.; Yu, D. Deep Learning: Methods and Applications. Found. Trends Signal Processing 2014, 7, 197–387.
- Nanni, L.; Brahnam, S.; Ghidoni, S.; Menegatti, E.; Barrier, T. A comparison of methods for extracting information from the co-occurrence matrix for subcellular classification. Expert Syst. Appl. 2013, 40, 7457–7467.
- Xu, Y.; Zhu, J.Y.; Eric, I.; Chang, C.; Lai, M.; Tu, Z. Weakly supervised histopathology cancer image segmentation and classification. Med. Image Anal. 2014, 18, 591–604.
- Barker, J.; Hoogi, A.; Depeursinge, A.; Rubin, D.L. Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med. Image Anal. 2015, 30, 60–71.
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000.
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117.
- Feng, C.; Elazab, A.; Yang, P.; Wang, T.; Zhou, F.; Hu, H.; Xiao, X.; Lei, B. Deep learning framework for alzheimer’s disease diagnosis via 3d-cnn and fsbi-lstm. IEEE Access 2019, 7, 63605–63618.
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444.
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the 27th International Conference on Neural Information Processing Systems, Ser. NIPS’14, Montreal, QC, Canada, 8–13 December 2014; MIT Press: Cambridge, MA, USA, 2014; Volume 2, pp. 3320–3328.
- Sarraf, S.; Tofighi, G. Classification of alzheimer’s disease using fmri data and deep learning convolutional neural networks. arXiv 2016, arXiv:1603.08631.
- Li, Y.; Huang, C.; Ding, L.; Li, Z.; Pan, Y.; Gao, X. Deep learning in bioinformatics: Introduction, application, and perspective in the big data era. Methods 2019, 166, 4–21.
- Mussap, M.; Noto, A.; Cibecchini, F.; Fanos, V. The importance of biomarkers in neonatology. Semin. Fetal Neonatal Med. 2013, 18, 56–64.
- Cedazo-Minguez, A.; Winblad, B. Biomarkers for Alzheimer’s disease and other forms of dementia: Clinical needs, limitations and future aspects. Exp. Gerontol. 2010, 45, 5–14.
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90.
- Shijie, J.; Ping, W.; Peiyi, J.; Siping, H. Research on data augmentation for image classification based on convolution neural networks. In Proceedings of the 2017 Chinese Automation Congress (CAC), Jinan, China, 20–22 October 2017; pp. 4165–4170.
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. Gan-based synthetic medical image augmentation for increased cnn performance in liver lesion classification. Neurocomputing 2018, 321, 321–331.
- Zhao, D.; Zhu, D.; Lu, J.; Luo, Y.; Zhang, G. Synthetic medical images using f&bgan for improved lung nodules classification by multi-scale vgg16. Symmetry 2018, 10, 519.
- Long, M.; Cao, Y.; Cao, Z.; Wnag, J.; Jordan, M.I. Transferable Representation Learning with Deep Adaptation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 3071–3085.
- Pichler, B.J.; Kolb, A.; Nägele, T.; Schlemmer, H.-P. PET/MRI: Paving the Way for the Next Generation of Clinical Multimodality Imaging Applications. J. Nucl. Med. 2010, 51, 333–336.
- Ding, J.; Chen, B.; Liu, H.; Huang, M. Convolutional neural network with data augmentation for sar target recognition. IEEE Geosci. Remote Sens. Lett. 2016, 13, 364–368.
- Castro, E.; Cardoso, J.S.; Pereira, J.C. Elastic deformations for data augmentation in breast cancer mass detection. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical Health Informatics (BHI), Las Vegas, NV, USA, 4–7 March 2018; pp. 230–234.
- Nordberg, A.; Rinne, J.O.; Kadir, A.; Långström, B. The use of PET in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 78–87.
- Shen, T.; Jiang, J.; Li, Y.; Wu, P.; Zuo, C.; Yan, Z. Decision supporting model for one-year conversion probability from mci to ad using cnn and svm. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 738–741.
- Shmulev, Y.; Belyaev, M. Predicting conversion of mild cognitive impairments to alzheimer’s disease and exploring impact of neuroimaging. In Graphs in Biomedical Image Analysis and Integrating Medical Imaging and Non-Imaging Modalities; Stoyanov, D., Taylor, Z., Ferrante, E., Dalca, A.V., Martel, A., Maier-Hein, L., Parisot, S., Sotiras, A., Papiez, B., Sabuncu, M.R., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 83–91.
- Suk, H.-I.; Lee, S.W.; Shen, D. Hierarchical feature representation and multimodal fusion with deep learning for ad/mci diagnosis. NeuroImage 2014, 101, 569–582.
- Nho, K.; Shen, L.; Kim, S.; Risacher, S.L.; West, J.D.; Foroud, T.; Jack, C.R., Jr.; Weiner, M.W.; Saykin, A.J. Automatic prediction of conversion from mild cognitive impairment to probable alzheimer’s disease using structural magnetic resonance imaging. In Annual Symposium Proceedings/AMIA Symposium; American Medical Informatics Association: Bethesda, MD, USA, 2010; Volume 2010, pp. 542–546.
- Costafreda, S.G.; Dinov, I.D.; Tu, Z.; Shi, Y.; Liu, C.Y.; Kloszewska, I.; Mecocci, P.; Soininen, H.; Tsolaki, M.; Vellas, B.; et al. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. NeuroImage 2011, 56, 212–219.
- Saraiva, C.; Praça, C.; Ferreira, R.; Santos, T.; Ferreira, L.; Bernardino, L. Nanoparticle-mediated brain drug delivery: Overcoming blood–brain barrier to treat neurodegenerative diseases. J. Control. Release 2016, 235, 34–47.
- Coupé, P.; Eskildsen, S.F.; Manjón, J.V.; Fonov, V.S.; Collins, D.L. Simultaneous segmentation and grading of anatomical structures for patient’s classification: Application to Alzheimer’s disease. Neuroimage 2011, 59, 3736–3747.
- Wolz, R.; Julkunen, V.; Koikkalainen, J.; Niskanen, E.; Zhang, D.P.; Rueckert, D.; Soininen, H.; Lötjönen, J.; The Alzheimer’s Disease Neuroimaging Initiative. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PLoS ONE 2011, 6, e25446.
- Liu, M.; Cheng, D.; Wang, K.; Wang, Y.; The Alzheimer’s Disease Neuroimaging Initiative. Multi-modality cascaded convolutional neural networks for Alzheimer’s disease diagnosis. Neuroinformatics 2018, 16, 295–308.
- Kruthika, K.R.; Maheshappa, H.D. Multistage classifier-based approach for Alzheimer’s disease prediction and retrieval. Inform. Med. Unlocked 2019, 14, 34–42.
- Basaia, S.; Agosta, F.; Wagner, L.; Canu, E.; Magnani, G.; Santangelo, R.; Filippi, M. Automated classification of Alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks. NeuroImage Clin. 2018, 21, 101645.
- Payan, A.; Montana, G. Predicting Alzheimer’s disease: A neuroimaging study with 3d convolutional neural networks. In Proceedings of the ICPRAM 2015 4th International Conference on Pattern Recognition Applications and Methods, Lisbon, Portugal, 10–12 January 2015; Volume 2.
- Asl, E.H.; Ghazal, M.; Mahmoud, A.; Aslantas, A.; Shalaby, A.; Casanova, M.; Barnes, G.; Gimel’farb, G.; Keynton, R.; El Baz, A. Alzheimer’s disease diagnostics by a 3d deeply supervised adaptable convolutional network. Front. Biosci. 2018, 23, 584–596.
- Feldman, M.D. Positron Emission Tomography (PET) for the Evaluation of Alzheimer’s Disease/Dementia. In Proceedings of the California Technology Assessment Forum, New York, NY, USA, June 2010.
- Bengio, Y. Learning deep architectures for AI. Found. Trends Mach. Learn. 2009, 2, 1–127.
- Ciregan, D.; Meier, U.; Schmidhuber, J. Multi-column deep neural networks for image classification. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 3642–3649.
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; ACM: Stateline, NV, USA, 2012; pp. 1097–1105.
- Farabet, C.; Couprie, C.; Najman, L.; Lecun, Y. Learning hierarchical features for scene labeling. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1915–1929.
- Hinton, G.; Deng, L.; Yu, D.; Dahl, G.; Mohamed, A.-R.; Jaitly, N.; Senior, A.; Vanhoucke, V.; Nguyen, P.; Sainath, T.N.; et al. Deep neural networks for acoustic modeling in speech recognition. IEEE Signal Process. Mag. 2012, 29, 82–97.
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems 26. Burges, C.J.C., Bottou, L., Welling, M., Ghahramani, Z., Weinberger, K.Q., Eds.; ACM: Stateline, NV, USA, 2013; pp. 3111–3119.
- Jo, T.; Nho, K.; Saykin, A.J. Deep Learning in Alzheimer’s Disease: Diagnostic Classification and Prognostic Prediction Using Neuroimaging Data. Front. Aging Neurosci. 2019, 11, 220.
- Boureau, Y.-L.; Ponce, J.; Lecun, Y. A theoretical analysis of feature pooling in visual recognition. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 111–118.
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comp. Vision 2015, 115, 211–252.
- Bengio, Y. Deep learning of representations: Looking forward. In Proceedings of the International Conference on Statistical Language and Speech Processing, First International Conference, SLSP 2013, Tarragona, Spain, 29–31 July 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–37.
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828.
- Yi, H.; Sun, S.; Duan, X.; Chen, Z. A study on Deep Neural Networks framework. In Proceedings of the 2016 IEEE Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC), Xi’an, China, 3–5 October 2016.
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507.
- Werbos, P.J. Backwards differentiation in AD and neural nets: Past links and new opportunities. In Automatic Differentiation: Applications, Theory, and Implementations; Bücker, H.M., Corliss, G., Hovland, P., Naumann, U., Norris, B., Eds.; Springer: New York, NY, USA, 2006; pp. 15–34.
