The Food and Drug Administration (FDA) Animal Rule was devised to facilitate approval of candidate vaccines and therapeutics using animal survival data when human efficacy studies are not practical or ethical. This regulatory pathway is critical for candidates against pathogens with high case fatality rates that prohibit human challenge trials, as well as candidates with low and sporadic incidences of outbreaks that make human field trials difficult. Important components of a vaccine development plan for Animal Rule licensure are the identification of an immune correlate of protection and immunobridging to humans. The relationship of vaccine-induced immune responses to survival after vaccination and challenge must be established in validated animal models and then used to infer predictive vaccine efficacy in humans via immunobridging.
- immune correlate
- immunobridging
- Animal Rule
- ELISA
- PsVNA
- binding
- neutralization
- animal model
- vaccine
- filovirus
1. Introduction
- “There is a reasonably well-understood pathophysiological mechanism of the toxicity of the substance and its prevention or substantial reduction by the product;
- The effect is demonstrated in more than one animal species expected to react with a response predictive for humans, unless the effect is demonstrated in a single animal species that represents a sufficiently well-characterized animal model for predicting the response in humans;
- The animal study endpoint is clearly related to the desired benefit in humans, generally the enhancement of survival or prevention of major morbidity; and
- The data or information on the kinetics and pharmacodynamics of the product or other relevant data or information, in animals and humans, allows selection of an effective dose in humans” [1] (Sec. 314.610). (In the case of vaccines, immune responses are the relevant parameter, as pharmacodynamics and kinetics are not applicable) [7].
2. Animal Models and Immune Correlates of Protection
3. Overview of Immunobridging
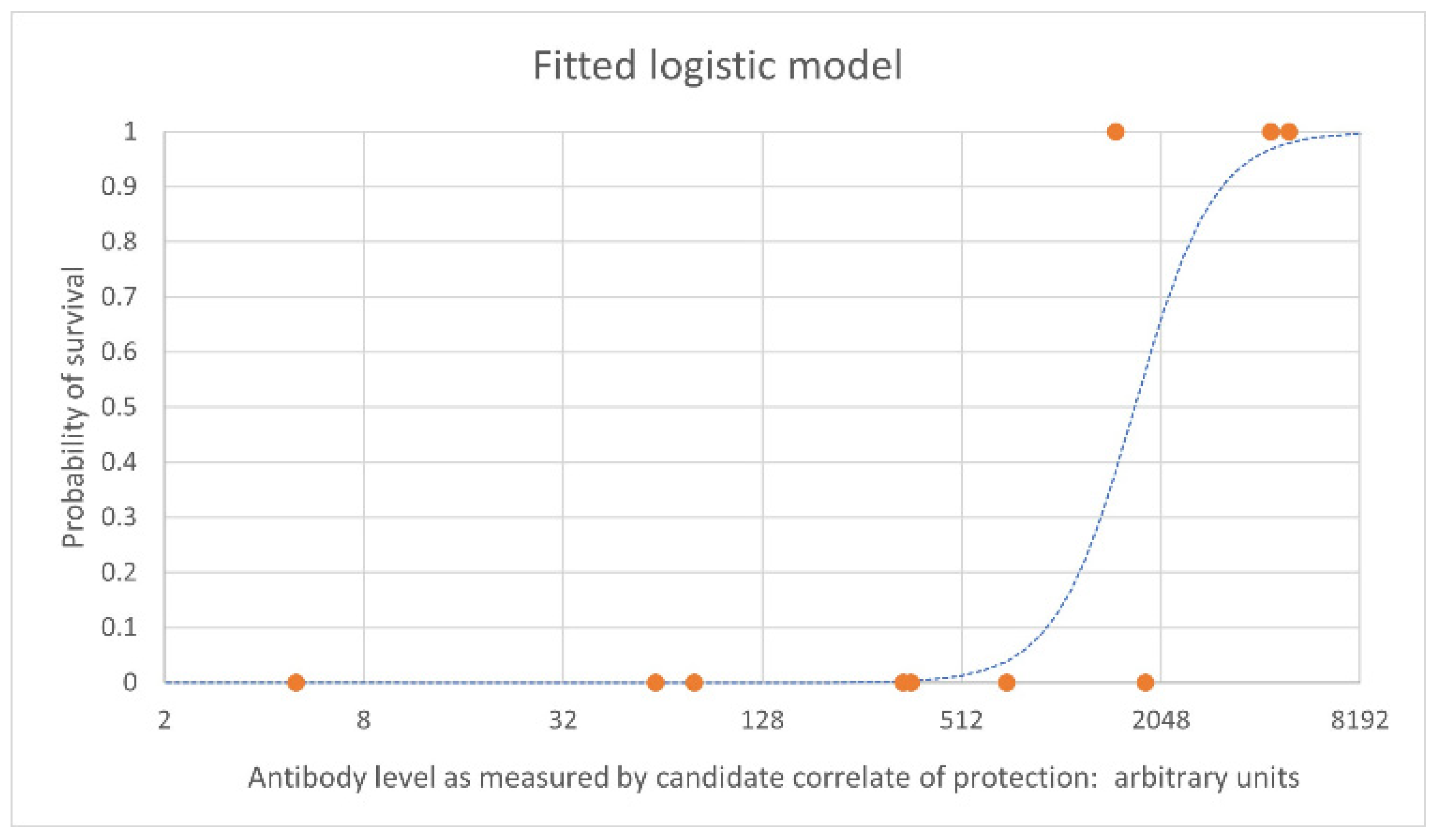
This entry is adapted from the peer-reviewed paper 10.3390/vaccines10091384
References
- U.S. Food and Drug Administration. Product Development under the Animal Rule Guidance for Industry. Available online: https://www.fda.gov/media/88625/download (accessed on 8 December 2021).
- Roozendaal, R.; Hendriks, J.; van Effelterre, T.; Spiessens, B.; Dekking, L.; Solforosi, L.; Czapska-Casey, D.; Bockstal, V.; Stoop, J.; Splinter, D.; et al. Nonhuman primate to human immunobridging to infer the protective effect of an Ebola virus vaccine candidate. NPJ Vaccines 2020, 5, 112.
- Ionin, B.; Hopkins, R.J.; Pleune, B.; Sivko, G.S.; Reid, F.M.; Clement, K.H.; Rudge, T.L.; Stark, G.V.; Innes, A.; Sari, S.; et al. Evaluation of immunogenicity and efficacy of anthrax vaccine adsorbed for postexposure prophylaxis. Clin. Vaccine Immunol. 2013, 20, 1016–1026.
- Krause, P.R.; Cavaleri, M.; Coleman, G.; Gruber, M.F. Approaches to demonstration of Ebola virus vaccine efficacy. Lancet Infect. Dis. 2015, 15, 627–629.
- European Medicines Agency. New Vaccine for Prevention of Ebola Virus Disease Recommended for Approval in the European Union. Available online: https://www.ema.europa.eu/en/news/new-vaccine-prevention-ebola-virus-disease-recommended-approval-european-union (accessed on 8 December 2021).
- Snoy, P.J. Establishing efficacy of human products using animals: The us food and drug administration’s “animal rule”. Vet. Pathol. 2010, 47, 774–778.
- Sullivan, N.J.; Martin, J.E.; Graham, B.S.; Nabel, G.J. Correlates of protective immunity for Ebola vaccines: Implications for regulatory approval by the animal rule. Nat. Rev. Microbiol. 2009, 7, 393–400.
- Kuhn, J.H.; Bao, Y.; Bavari, S.; Becker, S.; Bradfute, S.; Brister, J.R.; Bukreyev, A.A.; Caì, Y.; Chandran, K.; Davey, R.A.; et al. Virus nomenclature below the species level: A standardized nomenclature for laboratory animal-adapted strains and variants of viruses assigned to the family Filoviridae. Arch. Virol. 2013, 158, 1425–1432.
- Kuhn, J.H.; Adachi, T.; Adhikari, N.K.J.; Arribas, J.R.; Bah, I.E.; Bausch, D.G.; Bhadelia, N.; Borchert, M.; Brantsæter, A.B.; Brett-Major, D.M.; et al. New filovirus disease classification and nomenclature. Nat. Rev. Microbiol. 2019, 17, 261–263.
- Yang, X.L.; Tan, C.W.; Anderson, D.E.; Di Jiang, R.; Li, B.; Zhang, W.; Zhu, Y.; Lim, X.F.; Zhou, P.; Liu, X.L.; et al. Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat. Microbiol. 2019, 4, 390–395.
- Languon, S.; Quaye, O. Filovirus Disease Outbreaks: A Chronological Overview. Virol. Res. Treat. 2019, 10.
- Kuhn, J.H.; Amarasinghe, G.K.; Basler, C.F.; Bavari, S.; Bukreyev, A.; Chandran, K.; Crozier, I.; Dolnik, O.; Dye, J.M.; Formenty, P.B.H.; et al. ICTV virus taxonomy profile: Filoviridae. J. Gen. Virol. 2019, 100, 911–912.
- Interational Committee on Taxonomy of Viruses Filoviridae. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/w/filoviridae (accessed on 26 May 2022).
- Shi, M.; Lin, X.D.; Chen, X.; Tian, J.H.; Chen, L.J.; Li, K.; Wang, W.; Eden, J.S.; Shen, J.J.; Liu, L.; et al. The evolutionary history of vertebrate RNA viruses. Nature 2018, 556, 197–202.
- World Health Organization. Ebola Virus Disease. Available online: https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease (accessed on 8 January 2022).
- World Health Organization. Marburg Virus Disease. Available online: https://www.who.int/news-room/fact-sheets/detail/marburg-virus-disease (accessed on 8 January 2022).
- Feldmann, H.; Sprecher, A.; Geisbert, T.W. Ebola. N. Engl. J. Med. 2020, 382, 1832–1842.
- Kamorudeen, R.T.; Adedokun, K.A.; Olarinmoye, A.O. Ebola outbreak in West Africa, 2014–2016: Epidemic timeline, differential diagnoses, determining factors, and lessons for future response. J. Infect. Public Health 2020, 13, 956–962.
- Ledgerwood, J.E.; DeZure, A.D.; Stanley, D.A.; Coates, E.E.; Novik, L.; Enama, M.E.; Berkowitz, N.M.; Hu, Z.; Joshi, G.; Ploquin, A.; et al. Chimpanzee Adenovirus Vector Ebola Vaccine. N. Engl. J. Med. 2017, 376, 928–938.
- Albert, B. Sabin Vaccine Institute Evaluation of Safety, Tolerability and Immune Responses of Ebola-S and Marburg Vaccines in Healthy Adults. Available online: https://clinicaltrials.gov/ct2/show/NCT04723602?term=NCT04723602&draw=2&rank=1 (accessed on 8 January 2022).
- Suschak, J.J.; Schmaljohn, C.S. Vaccines against Ebola virus and Marburg virus: Recent advances and promising candidates. Hum. Vaccines Immunother. 2019, 15, 2359–2377.
- Lee, J.E.; Saphire, E.O. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009, 4, 2359–2377.
- Hoenen, T.; Groseth, A.; Feldmann, H. Therapeutic strategies to target the Ebola virus life cycle. Nat. Rev. Microbiol. 2019, 17, 593–606.
- Mulangu, S.; Dodd, L.E.; Davey, R.T.; Tshiani Mbaya, O.; Proschan, M.; Mukadi, D.; Lusakibanza Manzo, M.; Nzolo, D.; Tshomba Oloma, A.; Ibanda, A.; et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019, 381, 2293–2303.
- Wolf, J.; Jannat, R.; Dubey, S.; Troth, S.; Onorato, M.T.; Coller, B.A.; Hanson, M.E.; Simon, J.K. Development of pandemic vaccines: ERVEBO case study. Vaccines 2021, 9, 190.
- Hunegnaw, R.; Honko, A.; Wang, L.; Carr, D.; Murray, T.; Shi, W.; Dulan, C.N.M.; Foulds, K.E.; Agans, K.N.; Cross, R.W.; et al. Rapid and Durable Protection Against Marburg Virus with a Single-Shot ChAd3-MARV GP Vaccine. bioRxiv 2021.
- Corti, D.; Misasi, J.; Mulangu, S.; Stanley, D.A.; Kanekiyo, M.; Wollen, S.; Ploquin, A.; Doria-Rose, N.A.; Staupe, R.P.; Bailey, M.; et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 2016, 351, 1339–1342.
- U.S. Food and Drug Administration. FDA Approves Vaccine for Use after Known or Suspected Anthrax Exposure. Available online: https://wayback.archive-it.org/7993/20170111160749/http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm474027.htm (accessed on 27 March 2022).
- Marzi, A.; Banadyga, L.; Haddock, E.; Thomas, T.; Shen, K.; Horne, E.J.; Scott, D.P.; Feldmann, H.; Ebihara, H. A hamster model for Marburg virus infection accurately recapitulates Marburg hemorrhagic fever. Sci. Rep. 2016, 6, 39214.
- Bray, M.; Davis, K.; Geisbert, T.; Schmaljohn, C.; Huggins, J. A mouse model for evaluation of prophylaxis and therapy of ebola hemorrhagic fever. J. Infect. Dis. 1998, 178, 651–661.
- Connolly, B.M.; Steele, K.E.; Davis, K.J.; Geisbert, T.W.; Kell, W.M.; Jaax, N.K.; Jahrling, P.B. Pathogenesis of experimental Ebola virus infection in guinea pigs. Proc. J. Infect. Dis. 1999, 179, S203–S217.
- Raymond, J.; Bradfute, S.; Bray, M. Filovirus infection of STAT-1 knockout mice. J. Infect. Dis. 2011, 204, S986–S990.
- Atkins, C.; Miao, J.; Kalveram, B.; Juelich, T.; Smith, J.K.; Perez, D.; Zhang, L.; Westover, J.L.B.; Van Wettere, A.J.; Gowen, B.B.; et al. Natural History and Pathogenesis of Wild-Type Marburg Virus Infection in STAT2 Knockout Hamsters. Proc. J. Infect. Dis. 2018, 218, S438–S447.
- Escaffre, O.; Juelich, T.L.; Neef, N.; Massey, S.; Smith, J.; Brasel, T.; Smith, J.K.; Kalveram, B.; Zhang, L.; Perez, D.; et al. Stat-1 knockout mice as a model for wild-type sudan virus (Sudv). Viruses 2021, 13, 1388.
- Comer, J.E.; Escaffre, O.; Neef, N.; Brasel, T.; Juelich, T.L.; Smith, J.K.; Smith, J.; Kalveram, B.; Perez, D.D.; Massey, S.; et al. Filovirus virulence in interferon α/β and γ double knockout mice, and treatment with favipiravir. Viruses 2019, 11, 137.
- Cross, R.W.; Fenton, K.A.; Geisbert, J.B.; Ebihara, H.; Mire, C.E.; Geisbert, T.W. Comparison of the Pathogenesis of the Angola and Ravn Strains of Marburg Virus in the Outbred Guinea Pig Model. J. Infect. Dis. 2015, 212, S258–S270.
- Schiffman, Z.; Liu, G.; Cao, W.; Zhu, W.; Emeterio, K.; Qiu, X.; Banadyga, L. The Ferret as a Model for Filovirus Pathogenesis and Countermeasure Evaluation. ILAR J. 2020, 61, 62–71.
- Cross, R.W.; Mire, C.E.; Borisevich, V.; Geisbert, J.B.; Fenton, K.A.; Geisbert, T.W. The domestic ferret (mustela putorius furo) as a lethal infection model for 3 species of ebolavirus. J. Infect. Dis. 2016, 214, 565–569.
- Siragam, V.; Wong, G.; Qiu, X.G. Animal models for filovirus infections. Zool. Res. 2018, 39, 15.
- Bente, D.; Gren, J.; Strong, J.E.; Feldmann, H. Disease modeling for Ebola and Marburg viruses. DMM Dis. Model. Mech. 2009, 2, 12–17.
- Fasina, F.O.; Shittu, A.; Lazarus, D.; Tomori, O.; Simonsen, L.; Viboud, C.; Chowell, G. Transmission dynamics and control of ebola virus disease outbreak in Nigeria, July to september 2014. Eurosurveillance 2014, 19, 20920.
- Nakayama, E.; Saijo, M. Animal models for Ebola and Marburg virus infections. Front. Microbiol. 2013, 4, 267.
- St Claire, M.C.; Ragland, D.R.; Bollinger, L.; Jahrling, P.B. Animal models of Ebolavirus infection. Comp. Med. 2017, 67, 253–262.
- Glaze, E.R.; Roy, M.J.; Dalrymple, L.W.; Lanning, L.L. A comparison of the pathogenesis of marburg virus disease in humans and nonhuman primates and evaluation of the suitability of these animal models for predicting clinical efficacy under the ‘animal rule’. Comp. Med. 2015, 65, 241–259.
- Hérodin, F.; Thullier, P.; Garin, D.; Drouet, M. Nonhuman primates are relevant models for research in hematology, immunology and virology. Eur. Cytokine Netw. 2005, 16, 104–116.
- Kennedy, R.C.; Shearer, M.H.; Hildebrand, W. Nonhuman primate models to evaluate vaccine safety and immunogenicity. Proc. Vaccine 1997, 15, 903–908.
- Bradfute, S.B.; Warfield, K.L.; Bray, M. Mouse models for filovirus infections. Viruses 2012, 4, 1477–1508.
- Centers for Disease Control and Prevention. History of Marburg Virus Disease (MVD) Outbreaks. Available online: https://www.cdc.gov/vhf/marburg/outbreaks/chronology.html (accessed on 24 May 2022).
- Niemuth, N.A.; Fallacara, D.; Triplett, C.A.; Tamrakar, S.M.; Rajbhandari, A.; Florence, C.; Ward, L.; Griffiths, A.; Carrion, R.; Goez-Gazi, Y.; et al. Natural history of disease in cynomolgus monkeys exposed to Ebola virus Kikwit strain demonstrates the reliability of this nonhuman primate model for Ebola virus disease. PLoS ONE 2021, 16, e0252874.
- Alfson, K.J.; Goez-Gazi, Y.; Gazi, M.; Staples, H.; Mattix, M.; Ticer, A.; Klaffke, B.; Stanfield, K.; Escareno, P.; Keiser, P.; et al. Development of a well-characterized rhesus macaque model of ebola virus disease for support of product development. Microorganisms 2021, 9, 489.
- Blair, P.W.; Keshtkar-Jahromi, M.; Psoter, K.J.; Reisler, R.B.; Warren, T.K.; Johnston, S.C.; Goff, A.J.; Downey, L.G.; Bavari, S.; Cardile, A.P. Virulence of marburg virus Angola compared to Mt. Elgon (Musoke) in macaques: A pooled survival analysis. Viruses 2018, 10, 658.
- Herbert, A.S.; Froude, J.W.; Ortiz, R.A.; Kuehne, A.I.; Dorosky, D.E.; Bakken, R.R.; Zak, S.E.; Josleyn, N.M.; Musiychuk, K.; Mark Jones, R.; et al. Development of an antibody cocktail for treatment of Sudan virus infection. Proc. Natl. Acad. Sci. USA 2020, 117, 3768–3778.
- Ellis, D.S.; Bowen, E.T.W.; Simpson, D.I.H.; Stamford, S. Ebola virus: A comparison, at ultrastructural level, of the behaviour of the Sudan and Zaire strains in monkeys. Br. J. Exp. Pathol. 1978, 59, 584–593.
- Geisbert, T.W.; Daddario-DiCaprio, K.M.; Geisbert, J.B.; Reed, D.S.; Feldmann, F.; Grolla, A.; Ströher, U.; Fritz, E.A.; Hensley, L.E.; Jones, S.M.; et al. Vesicular stomatitis virus-based vaccines protect nonhuman primates against aerosol challenge with Ebola and Marburg viruses. Vaccine 2008, 26, 6894–6900.
- Henao-Restrepo, A.M.; Camacho, A.; Longini, I.M.; Watson, C.H.; Edmunds, W.J.; Egger, M.; Carroll, M.W.; Dean, N.E.; Diatta, I.; Doumbia, M.; et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: Final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!). Lancet 2017, 389, 505–518.
- Commission Report of an I. Ebola haemorrhagic fever in Zaire, 1976. Bull. World Health Organ. 1978, 56, 271–293.
- Beer, B.; Kurth, R.; Bukreyev, A. Characteristics of filoviridae: Marburg and Ebola viruses. Naturwissenschaften 1999, 86, 8–17.
- Meyer, M.; Malherbe, D.C.; Bukreyev, A. Can Ebola Virus Vaccines Have Universal Immune Correlates of protection? Trends Microbiol. 2019, 27, 8–16.
- Ilinykh, P.A.; Bukreyev, A. Antibody responses to filovirus infections in humans: Protective or not? Lancet Infect. Dis. 2021, 21, e348–e355.
- Bergwerk, M.; Gonen, T.; Lustig, Y.; Amit, S.; Lipsitch, M.; Cohen, C.; Mandelboim, M.; Levin, E.G.; Rubin, C.; Indenbaum, V.; et al. COVID-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021, 385, 1474–1484.
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211.
- Feng, S.; Phillips, D.J.; White, T.; Sayal, H.; Aley, P.K.; Bibi, S.; Dold, C.; Fuskova, M.; Gilbert, S.C.; Hirsch, I.; et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 2032–2040.
- Liu, S.; Moayeri, M.; Leppla, S.H. Anthrax lethal and edema toxins in anthrax pathogenesis. Trends Microbiol. 2014, 22, 317–325.
- Addetia, A.; Crawford, K.H.D.; Dingens, A.; Zhu, H.; Roychoudhury, P.; Huang, M.L.; Jerome, K.R.; Bloom, J.D.; Greninger, A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020, 58, e02107-20.
- Warfield, K.L.; Olinger, G.G. Protective role of cytotoxic T lymphocytes in filovirus hemorrhagic fever. J. Biomed. Biotechnol. 2011, 2011, 984241.
- Sullivan, N.J.; Hensley, L.; Asiedu, C.; Geisbert, T.W.; Stanley, D.; Johnson, J.; Honko, A.; Olinger, G.; Bailey, M.; Geisbert, J.B.; et al. CD8 + cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat. Med. 2011, 17, 1128–1131.
- Marzi, A.; Jankeel, A.; Menicucci, A.R.; Callison, J.; O’Donnell, K.L.; Feldmann, F.; Pinski, A.N.; Hanley, P.W.; Messaoudi, I. Single Dose of a VSV-Based Vaccine Rapidly Protects Macaques From Marburg Virus Disease. Front. Immunol. 2021, 12, 774026.
- Marzi, A.; Engelmann, F.; Feldmann, F.; Haberthur, K.; Shupert, W.L.; Brining, D.; Scott, D.P.; Geisbert, T.W.; Kawaoka, Y.; Katze, M.G.; et al. Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc. Natl. Acad. Sci. USA 2013, 110, 1893–1898.
- Stanley, D.A.; Honko, A.N.; Asiedu, C.; Trefry, J.C.; Lau-Kilby, A.W.; Johnson, J.C.; Hensley, L.; Ammendola, V.; Abbate, A.; Grazioli, F.; et al. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nat. Med. 2014, 20, 1126–1129.
- Wong, G.; Richardson, J.S.; Pillet, S.; Patel, A.; Qiu, X.; Alimonti, J.; Hogan, J.; Zhang, Y.; Takada, A.; Feldmann, H.; et al. Immune parameters correlate with protection against ebola virus infection in rodents and nonhuman primates. Sci. Transl. Med. 2012, 4, 158ra146.
- Bradfute, S.B.; Bavari, S. Correlates of immunity to filovirus infection. Viruses 2011, 3, 982–1000.
- Marzi, A.; Menicucci, A.R.; Engelmann, F.; Callison, J.; Horne, E.J.; Feldmann, F.; Jankeel, A.; Feldmann, H.; Messaoudi, I. Protection against marburg virus using a recombinant VSV-vaccine depends on T and B cell activation. Front. Immunol. 2019, 10, 3071.
- Woolsey, C.; Jankeel, A.; Matassov, D.; Geisbert, J.B.; Agans, K.N.; Borisevich, V.; Cross, R.W.; Deer, D.J.; Fenton, K.A.; Latham, T.E.; et al. Immune correlates of postexposure vaccine protection against Marburg virus. Sci. Rep. 2020, 10, 3071.
- Grais, R.F.; Kennedy, S.B.; Mahon, B.E.; Dubey, S.A.; Grant-Klein, R.J.; Liu, K.; Hartzel, J.; Coller, B.A.; Welebob, C.; Hanson, M.E.; et al. Estimation of the correlates of protection of the rVSVΔG-ZEBOV-GP Zaire ebolavirus vaccine: A post-hoc analysis of data from phase 2/3 clinical trials. Lancet Microbe 2021, 2, e70–e78.
- van Griensven, J.; Edwards, T.; de Lamballerie, X.; Semple, M.G.; Gallian, P.; Baize, S.; Horby, P.W.; Raoul, H.; Magassouba, N.; Antierens, A.; et al. Evaluation of Convalescent Plasma for Ebola Virus Disease in Guinea. N. Engl. J. Med. 2016, 374, 33–42.
- Dye, J.M.; Herbert, A.S.; Kuehne, A.I.; Barth, J.F.; Muhammad, M.A.; Zak, S.E.; Ortiz, R.A.; Prugar, L.I.; Pratt, W.D. Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc. Natl. Acad. Sci. USA 2012, 109, 5034–5039.
- Perdomo-Celis, F.; Salvato, M.S.; Medina-Moreno, S.; Zapata, J.C. T-cell response to viral hemorrhagic fevers. Vaccines 2019, 7, 11.
- Rimoin, A.W.; Lu, K.; Bramble, M.S.; Steffen, I.; Doshi, R.H.; Hoff, N.A.; Mukadi, P.; Nicholson, B.P.; Alfonso, V.H.; Olinger, G.; et al. Ebola Virus Neutralizing Antibodies Detectable in Survivors of theYambuku, Zaire Outbreak 40 Years after Infection. J. Infect. Dis. 2018, 217, 223–231.
- Bramble, M.S.; Hoff, N.; Gilchuk, P.; Mukadi, P.; Lu, K.; Doshi, R.H.; Steffen, I.; Nicholson, B.P.; Lipson, A.; Vashist, N.; et al. Pan-filovirus serum neutralizing antibodies in a subset of Congolese ebolavirus infection survivors. J. Infect. Dis. 2018, 218, 1929–1936.
- Sobarzo, A.; Groseth, A.; Dolnik, O.; Becker, S.; Lutwama, J.J.; Perelman, E.; Yavelsky, V.; Muhammad, M.; Kuehne, A.I.; Marks, R.S.; et al. Profile and persistence of the virus-specific neutralizing humoral immune response in human survivors of sudan ebolavirus (Gulu). J. Infect. Dis. 2013, 208, 299–309.
- Meyer, M.; Gunn, B.M.; Malherbe, D.C.; Gangavarapu, K.; Yoshida, A.; Pietzsch, C.; Kuzmina, N.A.; Saphire, E.O.; Collins, P.L.; Crowe, J.E.; et al. Ebola vaccine-induced protection in nonhuman primates correlates with antibody specificity and Fc-mediated effects. Sci. Transl. Med. 2021, 13, eabg6128.
- Warfield, K.L.; Howell, K.A.; Vu, H.; Geisbert, J.; Wong, G.; Shulenin, S.; Sproule, S.; Holtsberg, F.W.; Leung, D.W.; Amarasinghe, G.K.; et al. Role of Antibodies in Protection Against Ebola Virus in Nonhuman Primates Immunized with Three Vaccine Platforms. Proc. J. Infect. Dis. 2018, 218, S553–S564.
- Medaglini, D.; Santoro, F.; Siegrist, C.A. Correlates of vaccine-induced protective immunity against Ebola virus disease. Semin. Immunol. 2018, 39, 65–72.
- Tibshirani, B.E.R.J. An Introduction to the Bootstrap; Chapman and Hall: New York, NY, USA, 1993.
- Yellowlees, A.; Perry, R.H.J. Estimating vaccine efficacy using animal efficacy data. Eur. J. Pharmacol. 2015, 759, 63–68.
- Kohberger, R.C.; Jemiolo, D.; Noriega, F. Prediction of pertussis vaccine efficacy using a correlates of protection model. Vaccine 2008, 26, 3516–3521.
