Age estimation is a paramount issue in criminal, anthropological, and forensic research. Because of this, several areas of research have focused on the establishment of new approaches for age prediction, including bimolecular and anthropological methods. DNA methylation (DNAm) has arisen as one of the hottest topics in the field. Many studies have developed age-prediction models (APMs) based on evaluation of DNAm levels of many genes in different tissue types and using different methodological approaches. However, several challenges and confounder factors should be considered before using methylation levels for age estimation in forensic contexts.
- forensic science
- age estimation
- DNA methylation levels
- epigenetic age
1. Introduction
2. DNA Methylation (DNAm): An Epigenetic Mechanism
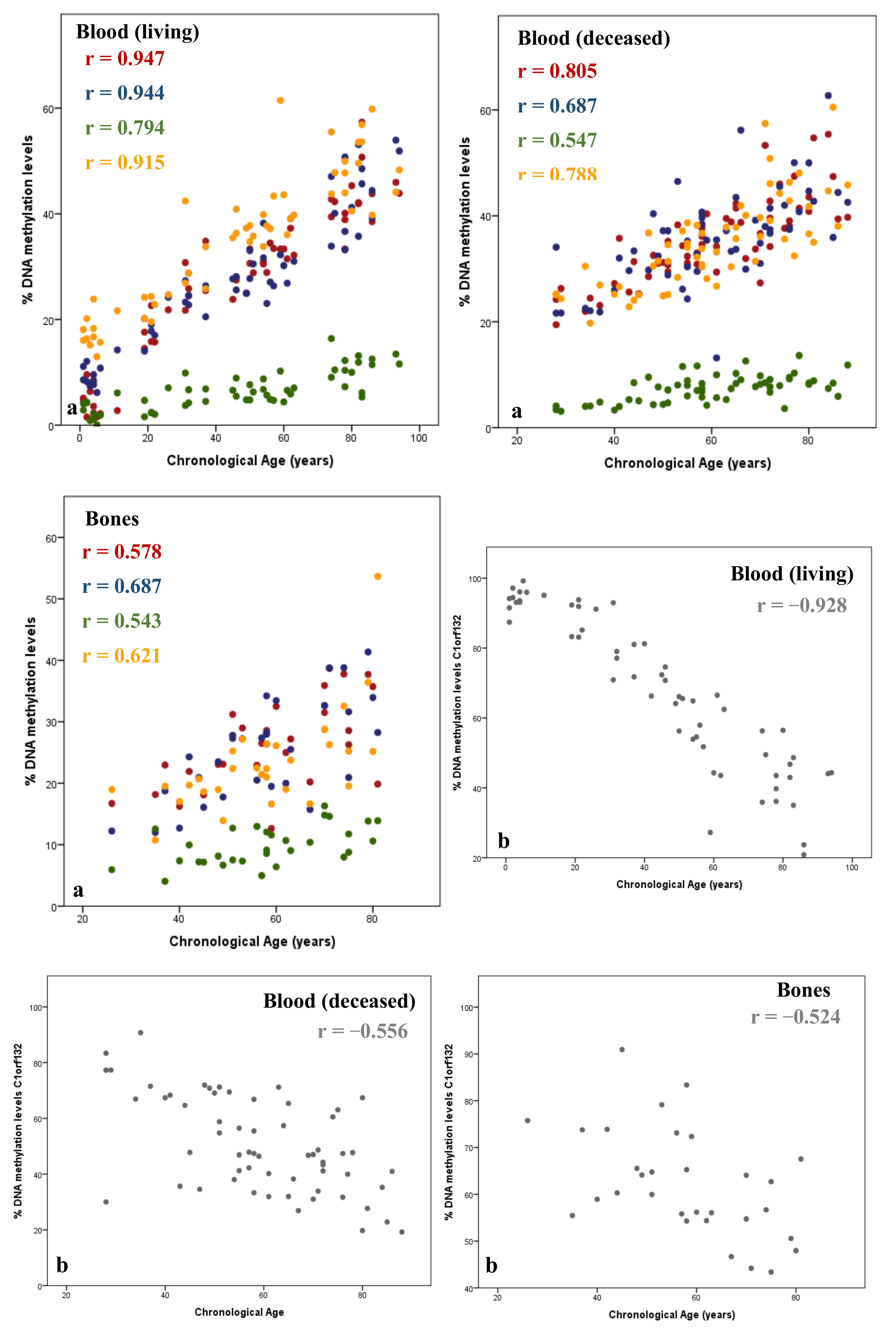
3. Underling Mechanisms of DNAm Changes with Age

4. Methodologies for DNAm Evaluation
This entry is adapted from the peer-reviewed paper 10.3390/forensicsci2030044
References
- Cunha, E.; Baccino, E.; Martrille, L.; Ramsthaler, F.; Prieto, J.; Schuliar, Y.; Lynnerup, N.; Cattaneo, C. The problem of aging human remains and living individuals: A review. Forensic Sci. Int. 2009, 193, 1–13.
- Franklin, D.; Flavel, A.; Noble, J.; Swift, L.; Karkhanis, S. Forensic age estimation in living individuals: Methodological considerations in the context of medico-legal practice. Res. Rep. Forensic Med. Sci. 2015, 5, 53–66.
- Parson, W. Age Estimation with DNA: From Forensic DNA Fingerprinting to Forensic (Epi) Genomics: A Mini-Review. Gerontology 2018, 64, 326–332.
- Nuzzolese, E.; Di Vella, G. Legal background of age estimation for the dead and the living. In Age Estimation: A Multidisciplinary Approach; Adserias-Garriga, J., Ed.; Elsevier: Oxford, UK, 2019; pp. 17–24.
- Cho, S.; Jung, S.E.; Hong, S.R.; Lee, E.H.; Lee, J.H.; Lee, S.D.; Lee, H.Y. Independent validation of DNA-based approaches for age prediction in blood. Forensic Sci. Int. Genet. 2017, 29, 250–256.
- Shi, L.; Jiang, F.; Ouyang, F.; Zhang, J.; Wang, Z.; Shen, X. DNA methylation markers in combination with skeletal and dental ages to improve age estimation in children. Forensic Sci. Int. Genet. 2018, 33, 1–9.
- Becker, J.; Mahlke, N.S.; Reckert, A.; Eickhoff, S.B.; Ritz-Timme, S. Age estimation based on different molecular clocks in several tissues and a multivariate approach: An explorative study. Int. J. Leg. Med. 2020, 134, 721–733.
- Baccino, E.; Cunha, E.; Cattaneo, C. Aging the Dead and the Living; Elsevier: Amsterdam, The Netherlands, 2013; pp. 42–48.
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115.
- Weidner, C.I.; Lin, Q.; Koch, C.M.; Eisele, L.; Beier, F.; Ziegler, P.; Bauerschlag, D.O.; Jöckel, K.H.; Erbel, R.; Mühleisen, T.W.; et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014, 15, R24.
- Huang, Y.; Yan, J.; Hou, J.; Fu, X.; Li, L.; Hou, Y. Developing a DNA methylation assay for human age prediction in blood and bloodstain. Forensic Sci. Int. Genet. 2015, 17, 129–136.
- Kader, F.; Ghai, M. DNA methylation and application in forensic sciences. Forensic Sci. Int. 2015, 249, 255–265.
- Lee, H.Y.; Lee, S.D.; Shin, K.-J. Forensic DNA methylation profiling from evidence material for investigative leads. BMB Rep. 2016, 49, 359–369.
- Zubakov, D.; Liu, F.; Kokmeijer, I.; Choi, Y.; van Meurs, J.B.J.; van Ijcken, W.F.J.; Uitterlinden, A.G.; Hofman, A.; Broer, L.; van Duijn, C.M.; et al. Human age estimation from blood using mRNA, DNA methylation, DNA rearrangement, and telomere length. Forensic Sci. Int. Genet. 2016, 24, 33–43.
- Jylhävä, J.; Pedersen, N.L.; Hägg, S. Biological age predictors. EBioMedicine 2017, 21, 29–36.
- Freire-Aradas, A.; Phillips, C.; Lareu, M.V. Forensic individual age estimation with DNA: From initial approaches to methylation tests. Forensic Sci. Rev. 2017, 29, 121–144.
- Vidaki, A.; Kayser, M. Recent progress, methods and perspectives in forensic epigenetics. Forensic Sci. Int. Genet. 2018, 37, 180–195.
- Williams, G. The emerging field of forensic epigenetics. Forensic Sci. Int. 2018, 290, e24–e25.
- Zolotarenko, A.D.; Chekalin, E.V.; Bruskin, S.A. Modern Molecular Genetic Methods for Age Estimation in Forensics. Russ J Genet. 2019, 55, 1460–1471.
- Hanafi, M.G.; Soedarsono, N.; Auerkari, E.I. Biological age estimation using DNA methylation analysis: A systematic review. Sci Dent. J. 2021, 5, 1–11.
- Roberti, A.; Valdes, A.F.; Torrecillas, R.; Fraga, M.F.; Fernandez, A.F. Epigenetics in cancer therapy and nanomedicine. Clin. Epigenetics 2019, 11, 81.
- Liu, X.; Jiao, B.; Shen, L. The Epigenetics of Alzheimer’s Disease: Factors and Therapeutic Implications. Front. Genet. 2018, 9, 579.
- Lardenoije, R.; Latrou, A.; Kenis, G.; Kompotis, K.; Steinbusch, H.W.M.; Mastroeni, D.; Coleman, P.; Lemere, C.A.; Hof, P.R.; van den Hove, D.L.A.; et al. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 2015, 131, 21–64.
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326.
- Jiang, S.; Guo, Y. Epigenetic Clock: DNA Methylation in Aging. Stem Cells Int. 2020, 2020, 1047896.
- Heyn, H.; Li, N.; Ferreira, H.J.; Moran, S.; Pisano, D.G.; Gomez, A.; Diez, J.; Sanchez-Mut, J.V.; Setien, F.; Javier Carmona, F.; et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl. Acad. Sci. USA 2012, 109, 10522–10527.
- Florath, I.; Butterbach, K.; Müller, H.; Bewerunge-Hudler, M.; Brenner, H. Cross-sectional and longitudinal changes in DNA methylation with age: An epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Human Mol. Genet. 2014, 23, 1186–1201.
- Zampieri, M.; Ciccarone, F.; Calabrese, R.; Franceschi, C.; Bürkled, A.; Caiafa, P. Reconfiguration of DNA methylation in aging. Mech. Ageing Dev. 2015, 151, 60–70.
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932.
- Xiao, F.H.; Kong, Q.P.; Perry, B.; He, Y.H. Progress on the role of DNA methylation in aging and longevity. Brief Funct. Genom. 2016, 15, 454–459.
- Garagnani, P.; Bacalini, M.G.; Pirazzini, C.; Gori, D.; Giuliani, C.; Mari, D.; Di Blasio, A.M.D.; Gentilini, D.; Vitale, G.; Collino, S.; et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell 2012, 11, 1132–1134.
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367.
- Bocklandt, S.; Lin, W.; Sehl, M.E.; Sanchez, F.J.; Sinsheimer, J.S.; Horvath, S.; Vilain, E. Epigenetic predictor of age. PLoS ONE 2011, 6, e14821.
- Pal, S.; Tyler, J.K. Epigenetics and aging. Sci. Adv. 2016, 2, e1600584.
- Fraga, M.F.; Ballestar, E.; Paz, M.F.; Ropero, S.; Setien, F.; Ballestar, M.L.; Heine-Suner, D.; Cigudosa, J.C.; Urioste, M.; Benitez, J.; et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA. 2005, 102, 10604–10609.
- Tan, Q.; Heijmans, B.T.; Hjelmborg, J.V.; Soerensen, M.; Christensen, K.; Christiansen, L. Epigenetic drift in the aging genome: A ten-year follow-up in an elderly twin cohort. Int. J. Epidemiol. 2016, 45, 1146–1158.
- Hasnain, S.E. Forensic Epigenetic Analysis: The Path Ahead. Med. Princ. Pract. 2019, 28, 301–308.
- Goel, N.; Karira, P.; Garg, V.K. Role of DNA methylation in human age prediction. Mech. Ageing Dev. 2017, 166, 33–41.
- Bergsma, T.; Rogaeva, E. DNA Methylation Clocks and Their Predictive Capacity for Aging Phenotypes and Healthspan. Neurosci. Insights 2020, 21, 2633105520942221.
- Correia Dias, H.; Cordeiro, C.; Pereira, J.; Pinto, C.; Corte Real, F.; Cunha, E.; Manco, L. DNA methylation age estimation in blood samples of living and deceased individuals using a multiplex SNaPshot assay. Forensic Sci. Int. 2020, 311, 110267.
- Dias, M.H.C. DNA methylation as an age predictor in living and deceased individuals. Ph.D. Thesis, Universidade de Coimbra, Coimbra, Portugal, 2021.
- Jung, S.E.; Lim, S.M.; Hong, S.R.; Lee, E.H.; Shin, K.J.; Lee, H.Y. DNA methylation of the ELOVL2, FHL2, KLF14, C1orf132/MIR29B2C, and TRIM59 genes for age prediction from blood, saliva, and buccal swab samples. Forensic Sci. Int. Genet. 2019, 38, 1–8.
- Eipel, M.; Mayer, F.; Arent, T.; Ferreira, M.R.P.; Birkhofer, C.; Gerstenmaier, U.; Costa, I.G.; Ritz-Timme, S.; Wagner, W. Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures. Aging 2016, 8, 1034–1048.
- Naue, J.; Sänger, T.; Hoefsloot, H.C.J.; Lutz-Bonengel, S.; Kloosterman, A.D.; Verschure, P.J. Proof of concept study of age-dependent DNA methylation markers across different tissues by massive parallel sequencing. Forensic Sci. Int. Genet. 2018, 36, 152–159.
- Correia Dias, H.; Manco, L.; Corte Real, F.; Cunha, E. A Blood-Bone-Tooth Model for Age Prediction in Forensic Contexts. Biology 2021, 10, 1312.
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384.
- Liu, Z.; Leung, D.; Levine, M. Comparative analysis of epigenetic aging clocks from CpG characteristics to functional Associations. BioRxiv 2019, 512483.
- Freire-Aradas, A.; Pośpiech, E.; Aliferi, A.; Girón-Santamaría, L.; Mosquera-Miguel, A.; Pisarek, A.; Ambroa-Conde, A.; Phillips, C.; Casares de Cal, M.A.; Gómez-Tato, A.; et al. A Comparison of Forensic Age Prediction Models Using Data From Four DNA Methylation Technologies. Front. Genet. 2020, 19, 932.
- Jiang, M.; Zhang, Y.; Fei, J.; Chang, X.; Fan, W.; Qian, X.; Zhang, T.; Lu, D. Rapid quantification of DNA methylation by measuring relative peak heights in direct bisulfite-PCR sequencing traces. Lab. Investig. 2010, 90, 282–290.
- Parrish, R.R.; Day, J.J.; Lubin, F.D. Direct bisulfite sequencing for examination of DNA methylation patterns with gene and nucleotide resolution from brain tissues. Curr. Protoc. Neurosci. 2012, 60, 7–24.
- Bekaert, B.; Kamalandua, A.; Zapico, S.C.; Van de Voorde, W.; Decorte, R. Improved age determination of blood and teeth samples using a selected set of DNA methylation markers. Epigenetics 2015, 10, 922–930.
- Bekaert, B.; Kamalandua, A.; Zapico, S.C.; Van de Voorde, W.; Decorte, R. A selective set of DNA-methylation markers for age determination of blood, teeth and buccal samples. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e144–e145.
- Zbieć-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Makowska, Ż.; Spas, A.; Parys-Proszek, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci. Int. Genet. 2015, 14, 161–167.
- Zbieć-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Parys-Proszek, A.; Makowska, Ż.; Pałeczka, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Development of a forensically useful age prediction method based on DNA methylation analysis. Forensic Sci. Int. Genet. 2015, 17, 173–179.
- Park, J.L.; Kim, J.H.; Seo, E.; Bae, D.H.; Kim, S.Y.; Lee, H.C.; Woo, K.M.; Kim, Y.S. Identification and evaluation of age-correlated DNA methylation markers for forensic use. Forensic Sci. Int. Genet. 2016, 23, 64–70.
- Thong, Z.; Chan, X.L.S.; Tan, J.Y.Y.; Loo, E.S.; Syn, C.K.C. Evaluation of DNA methylation-based age prediction on blood. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e249–e251.
- Spólnicka, M.; Pośpiech, E.; Pepłońska, B.; Zbieć-Piekarska, R.; Makowska, Ż.; Pięta, A.; Karłowska-Pik, J.; Ziemkiewicz, B.; Wężyk, M.; Gasperowicz, P.; et al. DNA methylation in ELOVL2 and C1orf132 correctly predicted chronological age of individuals from three disease groups. Int. J. Leg. Med. 2017, 132, 1–11.
- Daunay, A.; Baudrin, L.G.; Deleuze, J.F.; How-Kit, A. Evaluation of six blood-based age prediction models using DNA methylation analysis by pyrosequencing. Sci. Rep. 2019, 9, 8862.
- Pfeifer, M.; Bajanowski, T.; Helmus, J.; Poetsch, M. Inter-laboratory adaption of age estimation models by DNA methylation analysis-problems and solutions. Int. J. Leg. Med. 2020, 134, 953–961.
- Lee, H.Y.; Jung, S.-E.; Oh, Y.N.; Choi, A.; Yang, W.I.; Shin, K.-J. Epigenetic age signatures in the forensically relevant body fluid of semen: A preliminary study. Forensic Sci. Int. Genet. 2015, 19, 28–34.
- Hong, S.R.; Jung, S.E.; Lee, E.H.; Shin, K.J.; Yang, W.I.; Lee, H.Y. DNA methylation based age prediction from saliva: High age predictability by combination of 7 CpG markers. Forensic Sci. Int. Genet. 2017, 29, 118–125.
- Hong, S.R.; Shin, K.-J.; Jung, S.-E.; Lee, E.H.; Lee, H.Y. Platform-independent models for age prediction using DNA methylation data. Forensic Sci. Int. Genet. 2019, 38, 39–47.
- Han, Y.; Franzen, J.; Stiehl, T.; Gobs, M.; Kuo, C.C.; Nikolić, M.; Hapala, J.; Koop, B.E.; Strathmann, K.; Ritz-Timme, S.; et al. New targeted approaches for epigenetic age predictions. BMC Biol. 2020, 18, 71.
- Manco, L.; Dias, H.C. DNA methylation analysis of ELOVL2 gene using droplet digital PCR for age estimation purposes. Forensic Sci. Int. 2022, 333, 111206.
- Woźniak, A.; Heidegger, A.; Piniewska-Róg, D.; Pośpiech, E.; Xavier, C.; Pisarek, A.; Kartasińska, E.; Boroń, M.; Freire-Aradas, A.; Wojtas, M.; et al. Development of the VISAGE enhanced tool and statistical models for epigenetic age estimation in blood, buccal cells and bones. Aging 2021, 11, 6459–6484.
- Vidaki, A.; Ballard, D.; Aliferi, A.; Miller, T.H.; Barron, L.P.; Syndercombe Court, D. DNA methylation-based forensic age prediction using artificial neural networks and next generation sequencing. Forensic Sci. Int. Genet. 2017, 28, 225–236.
- Aliferi, A.; Ballard, D.; Gallidabino, M.D.; Thurtle, H.; Barron, L.; Syndercombe Court, D. DNA methylation-based age prediction using massively parallel sequencing data and multiple machine learning models. Forensic Sci. Int. Genet. 2018, 37, 215–226.
- Naue, J.; Hoefsloot, H.C.J.; Mook, O.R.F.; Rijlaarsdam-Hoekstra, L.; van der Zwalm, M.C.H.; Henneman, P.; Kloosterman, A.D.; Verschure, P.J. Chronological age prediction based on DNA methylation: Massive parallel sequencing and random forest regression. Forensic Sci. Int. Genet. 2017, 31, 19–28.
