Metastasis detection in lymph nodes via microscopic examination of histopathological images is one of the most crucial diagnostic procedures for breast cancer staging. The manual analysis is extremely labor-intensive and time-consuming because of complexities and diversities of histopathology images. Deep learning has been utilized in automatic cancer metastasis detection in recent years. Due to the huge size of whole-slide images, most existing approaches split each image into smaller patches and simply treat these patches independently, which ignores the spatial correlations among them. To solve this problem, this paper proposes an effective spatially sensitive learning framework for cancer metastasis detection in whole-slide images. Moreover, a novel spatial loss function is designed to ensure the consistency of prediction over neighboring patches. Specifically, through incorporating long short-term memory and spatial loss constraint on top of a convolutional
neural network feature extractor, the proposed method can effectively learn both the appearance of each patch and spatial relationships between adjacent image patches. With the standard backpropagation algorithm, the whole framework can be trained in an end-to-end way. Finally, the regions with high tumor probability in the resulting probability map are the metastasis locations. Extensive experiments on the benchmark Camelyon 2016 Grand Challenge dataset show the effectiveness of
the proposed approach with respect to state-of-the-art competitors. The obtained precision, recall, and balanced accuracy are 0.9565, 0.9167, and 0.9458, respectively. It is also demonstrated that the proposed approach can provide more accurate detection results and is helpful for early diagnosis of cancer metastasis.
1. Introduction
Cancer is currently one of the major causes of death for people all over the world. It is estimated that 14.5 million people have died of cancer, and by 2030, this figure is expected to exceed 28 million. The most common cancer in women is breast cancer. Every year, 2.1 million people around the world are diagnosed with breast cancer according to the World Health Organization (WHO) [
1]. Due to the high rate of mortality, considerable efforts have been made in recent decades to detect breast cancer from histological images so as to improve survival through early breast cancer detection or through breast tissue diagnosis.
Because lymph nodes are the first site of breast cancer metastasis, the metastasis identification of lymph nodes is one of the most essential criteria for early detection [
2]. In order to analyze the characteristics of tissues, pathologists examine the tissue slices under a microscope [
3]. The tissue slices are traditionally directly observed with the histopathologist’s naked eye, and visual data are assessed manually based on prior medical knowledge. The manual analysis is highly time consuming and labor intensive due to the diversities of histopathological images, especially for tiny lymph nodes. At the same time, depending on the histopathologist’s expertise, workload, and current mood, the manual diagnostic procedure is subjective and has limited repeatability. In addition, in the face of escalating diagnostic demands with increased cancer incidence, there is a serious shortage of pathologists [
4]. Hundreds of biopsies must be diagnosed daily by pathologists, thus it is almost impossible to thoroughly examine entire slides. However, if only regions of interest are investigated, the chance of incorrect diagnosis may increase. To this end, in order to increase the efficiency and reliability of pathological examination, it is required to develop automatic detection techniques. Therefore, computer aided diagnosis (CAD) is established to improve the consistency, efficiency, and sensitivity of metastasis identification [
5].
However, automated metastasis identification in sentinel lymph nodes from a whole-slide image (WSI) is extremely challenging for the following reasons. First, hard imitations in normal tissues usually look similar in morphology to metastatic areas, which leads to many false positives. Second, there is a great variety of biological structures and textures of metastatic and background areas. Third, the varied circumstances of histological image processing (for example, staining, cutting, sampling, and digitization) enhance the variations of the appearance of the image. This usually happens as tissue samples are taken at different time points or from different patients. Last but not least, WSIs are incredibly huge, approximately 100,000 pixels × 200,000 pixels, and may not be directly input into any existing methods for cancer identification. Another problem for automatic detection algorithms is how to analyze such a large pixel image effectively.
Artificial intelligence (AI) technologies have developed rapidly in recent years and achieve outstanding breakthroughs, especially in computer vision, image processing and analysis. In histopathological diagnosis, AI has also exhibited potential advantages. With the help of AI-assisted diagnostic approaches, valuable information about diagnostics may be speedily extracted from big data, alleviating the workload of pathologists. At the same time, AI-aided diagnostics have more objective analysis capabilities and can avoid subjective discrepancies of manual analysis. To a certain extent, artificial intelligence helps not only to improve work efficiency but also to reduce the rate of misdiagnosis by pathologists.
In the past few decades, much work on breast histology image recognition has been done. Early research used hand-made features to capture tissue properties in specific areas for automatic detection [
6,
7,
8]. However, hand-made features are not sufficiently discriminative to describe a wide variety of shapes and textures. With the emergence of powerful computers, deep learning technology has made remarkable progress in a variety of domains, including natural language understanding, speech recognition, computer vision, and image processing [
9]. These methods have also been successfully employed in various modalities of medical images for classification, detection, and segmentation tasks [
10,
11,
12]. Recently, deep convolutional neural networks (CNNs) have been utilized to detect cancer metastases that can learn more effective feature representation and obtain higher detection accuracy in a data-driven approach [
13,
14,
15]. As the size of WSI is extremely huge, most studies first extracted tiny patches (for example, 256 pixels × 256 pixels) from WSI, then deep CNN was trained to categorize these tiny patches to be tumorous or normal. The probability map was subsequently produced at the patch level and used for the metastasis identification of the original WSI. The spatial relationships were not explicitly modeled, as the patches were independently extracted and trained. Consequently, the predictions from adjacent regions may not be consistent in the inference stage.
The main contributions of this paper are:
• This paper proposes a new spatially sensitive learning architecture that integrates CNN and long short-term memory (LSTM) in a unified framework to automatically
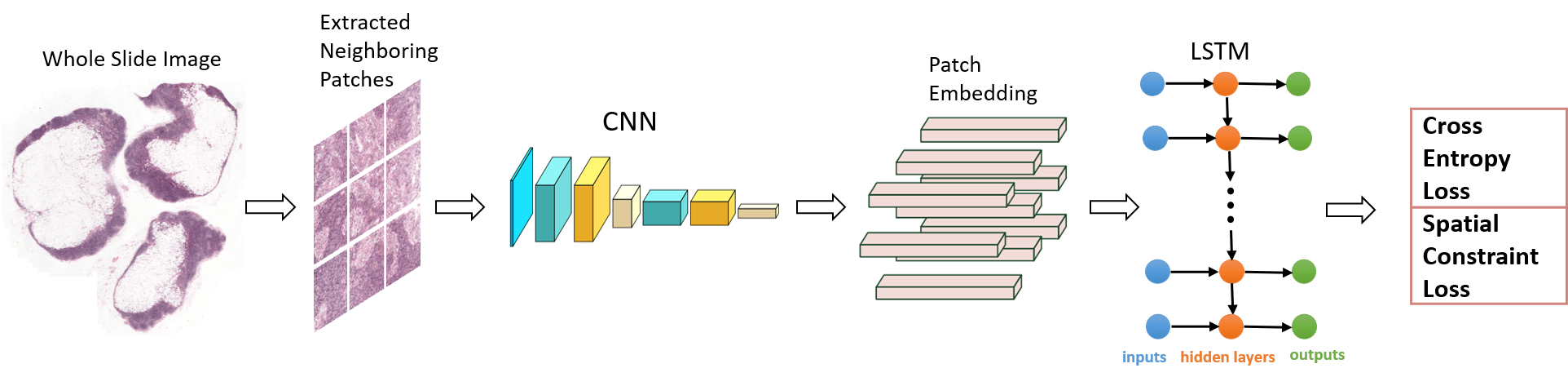
detect the metastasis locations, as shown in Figure 1.
• Inspired by the observation that adjacent regions are interrelated, an LSTM layer is employed to explicitly describe the spatial correlation, at the same time, spatial
constraint is also imposed on the loss function to further improve performance.
• Unlike previous approaches, the proposed model not only takes into account the appearance of each patch, but the spatial information between adjacent areas is also embedded into the framework to make better predictions.
Figure 1. Overview of the proposed framework. CNN and LSTM are two important components of this framework. When a grid of patches is input, the CNN feature extractor will encode every patch as a given-length vector representation. Given the grid of patch representations, the LSTM component models the spatial relationships between adjacent patches and outputs the probability of every patch being tumorous. In addition, spatial constraint is also imposed on the loss function to further improve the performance.
2. Related Work
In earlier years, most approaches employed hand-crafted features for cancer metastasis detection. In Reference [
6], authors distinguished the malignant from the benign based on several hand-crafted textural features. Some studies merged two or more hand-crafted features to enhance the accuracy of detection. In Reference [
7], graph, haralick, local binary patterns (LBP), and intensity features were used for cancer identification of H&E stained histopathological images. In Reference [
8], histopathological images were represented via fusing color histograms, LBP, SIFT, and some kernel features, and the significance of these pattern features was also studied. However, it takes considerable effort to design and validate the hand-made features. In addition, the properties of tissues with great variations in morphology and texture cannot properly be represented. Therefore, the detection performance of these methods based on hand-crafted features is poor.
In Reference [
16], authors developed a deep learning system to study the stromal properties of breast tissues associated with tumor for classifying whole slide images (WSIs). Authors in Reference [
13] utilized AlexNet to categorize breast cancer in histopathological images to be malignant or benign. Authors in Reference [
14] developed two distinct CNN architectures to classify breast cancer of pathology images. Single-task CNN was applied to identify malignant tumors. Multi-task CNN was used for analyzing the properties of benign and malignant tumors. The hybrid CNN unit designed in Reference [
15] could fully exploit the global and local features of images, and thus obtained superior prediction performance. In References [
17,
18], authors proposed a dense and fast screening architecture (ScanNet) to identify metastatic breast cancer in WSIs. Several methods based on transfer learning are also proposed to detect breast cancer [
19,
20,
21]. However, these above approaches deal with each patch of an image individually. In reality, each image patch and its adjacent ones generally share spatial correlations that are crucial for prediction. If one patch is a tumor, its adjacent patches are more likely in the tumor region, as they are situated in the neighboring areas.
In order to fully capture the spatial structure information between adjacent patches, authors in Reference [
22] applied a conditional random field (CRF) on patch features, which are first obtained from a CNN classifier. However, the method [
22] adopted a two-step learning framework, so the spatial relationships between neighboring patches are not available for the CNN feature extractor. Authors in Reference [
23] employed 2D long short-term memory (LSTM) on patch features to learn spatial structure information, due to higher computation cost of the end-to-end learning scheme, the two-stage learning strategy was also adopted in their experiments [
23].
This paper presents an alternative spatially sensitive learning architecture to fully explore spatial relationships between neighboring patches. This model is composed of a CNN feature extractor, LSTM, and spatial loss constraint. The standard back-propagation algorithm can be utilized to train this architecture in an end-to-end way, and a postprocessing step is no longer required. As a result, the spatial structure information between adjacent patches is available for the CNN feature extractor, which can benefit from joint learning with the spatial constraint components.
3. Method
The proposed spatially sensitive learning method is introduced in this section. Its overall framework is shown in Figure 1. CNN and LSTM are two important components of this framework. When a grid of patches is input, the CNN feature extractor will encode every patch as a given-length vector representation. Given the grid of patch representations, the LSTM component models the spatial relationships between adjacent patches and outputs the probability of every patch being tumorous. The details of each component are presented in the next subsections.
This entry is adapted from the peer-reviewed paper 10.3390/math10152657