Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Research relating to dielectrophoresis (DEP) has been progressing rapidly through time as it is a strong and controllable technique for manipulation, separation, preconcentration, and partitioning of protein. Extensive studies have been carried out on protein DEP, especially on Bovine Serum Albumin (BSA).
- dielectrophoresis
- protein
- albumin
1. Theory of DEP
Dielectrophoresis (DEP) theory was discovered by Herbert Pohl in the 1950s when the sample used was on liquidated particles [1][2]. In his paper, he refers to the DEP response as ‘dielectro-precipitation’, which is when the polarizability of the liquidated particles is bigger than the solvent, causing the concentrated suspension to be more susceptible to collisions or congelation. DEP theory has been further studied in terms of the manipulation of bioparticles. In a recent study, research on DEP is implemented in dialyzers of kidney hemodialysis [3][4].
DEP allows concentrating, transferring, capturing, refinement, and enhancement of biological or clinical samples [5][6]. There are two types of DEP responses that highly rely on the conductivity and permittivity of the suspending medium and the bioparticles [7]. pDEP, also known as positive-DEP, is when the particles exerted with force are seen to be attracted to the edge of the electrodes (regions with high electric field). nDEP, also known as negative-DEP, is when the particles exerted with force are seen to be repelled towards the center of the electrodes (regions with low electric field). These DEP responses, as shown in Figure 1a–c, are expected of protein when being manipulated with DEP [8].
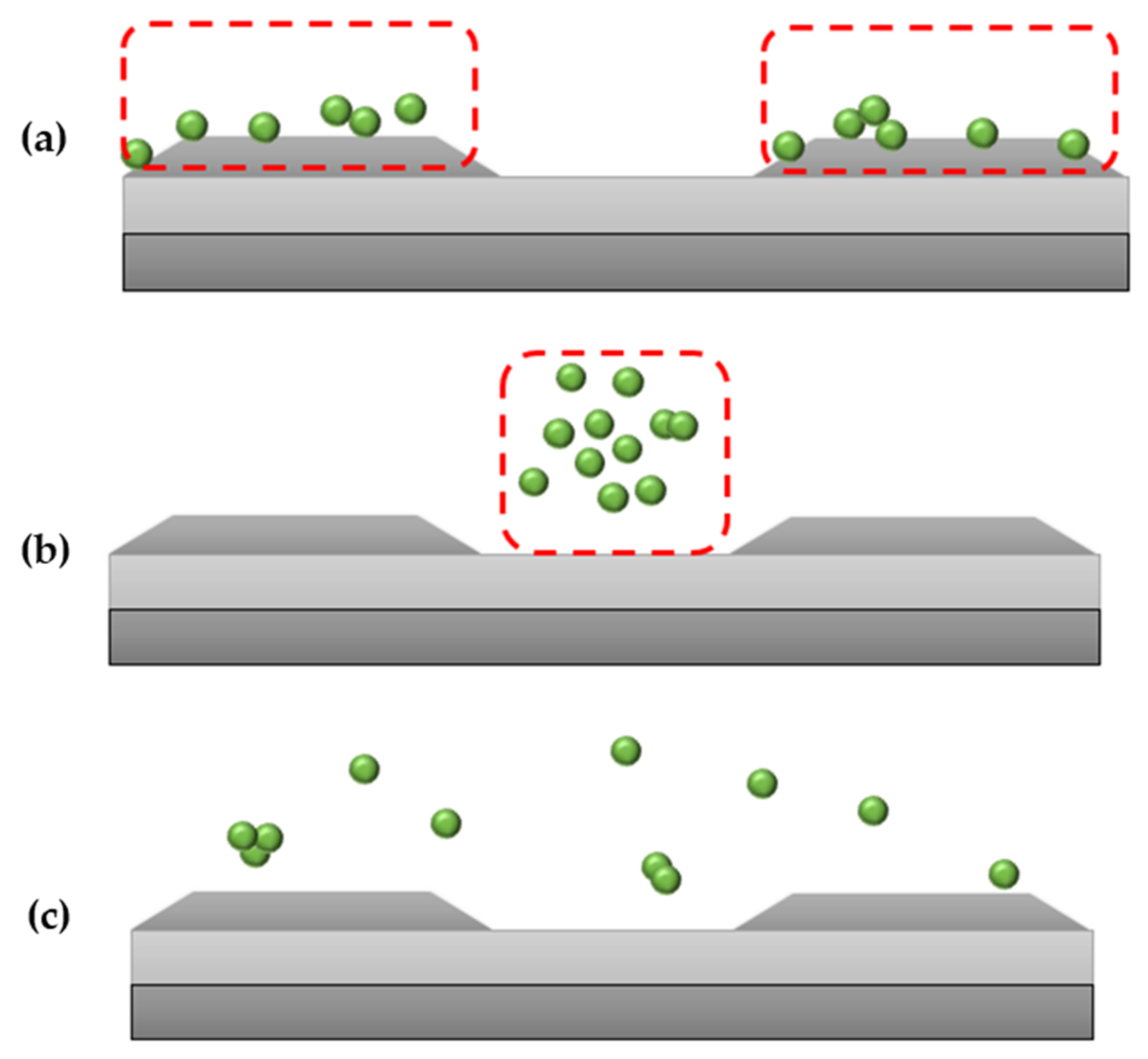
Figure 1. (a) pDEP or positive-DEP where particles are seen to be attracted to the edge of the electrodes. Particles move to the area with a higher field gradient, (b) nDEP or negative-DEP, where particles are seen repelled towards the center of the electrodes. Particles move away from the area with a high field gradient, (c) also known as crossover frequency, where particles can be seen dispersed around the electrodes.
In DEP, the particles subjected to manipulation can be applied with either alternating current (AC) or direct current (DC) nonuniform electric field [9]. However, ideally, AC is more suitable for DEP because DC tends to give rise to Faradaic reaction due to electrode-electrolyte interaction causing the formation of bubbles and other undesired responses [10]. The time-averaged DEP force ( ) exerted on spherical particles with radius, r can be calculated as in the equations below [11][12][13][14].
εp represents the permittivity of the particle while εm is the permittivity of the medium. is the complex permittivity which can be calculated in Equation (6). σ is the conductivity, and ω is the angular frequency. is the vacuum permittivity, where the value is 8.854 × 10−12 F/m [5]. CMf is the Clausius-Mossotti factor, also known as the Maxwell-Wagner factor. CMf contains two parts which are the real part and the imaginary part. The real part of CMf dictates the force in the direction of the highest or lowest point of an electric field. pDEP and nDEP responses can be determined by CMf.
The CMf is at its highest during pDEP and lowest during nDEP [15]. When the particle is seen to be more polarizable than the suspending medium (Re(CMf) > 0), the DEP response will be pDEP. When the particle is seen to be less polarizable than the suspending medium (Re(CMf) < 0), the DEP response will be nDEP [16]. Protein albumin is known to be ellipsoidal [17]. The equation to calculate exerted on ellipsoidal particle can be calculated as below:
where stands for dimension on the x-axis, b is on the y-axis, while c is on the z-axis [18]. CMf can be calculated as the equation below:
α stands for x,y or the z-axis while Aα is the depolarization factor. Based on the paper written by [19], proteins have a permanent dipole moment due to their interaction with water hydration forming hydration sheath and the total charges interaction within the protein cavity [20]. This is different from other biomacromolecules such as cells, bacteria, virions and up to vesicles that possess an induced moment. A dipole moment can be defined as the separation of charge within a region.
For a normal induced dipole moment model of DEP exerted on a spherical particle, the real part of the CMf value ranges from 1.0 > CMf > −0.5 [21]. According to Pethig, the dipole moment affects DEP response [19]. Equation (10) below is described to explain the of a molecular structure of spherical E. coli rRNA, which is a form of protein:
μp is the permanent dipole moment while μind is the induced dipole moment. A is the polarizability, and v is the volume. The symbol ∇ Eo could contribute to a pDEP response. ∇ plays an important role in an nDEP response. For manipulation of protein using DEP, the DEP force is exerted by the surrounding suspending medium of the protein. However, this equation is not applicable for globular protein. This shows that the equation for calculating DEP force is different for calculations of protein from the standard force of DEP equation.
In 2022, Pethig further developed an equation for protein DEP with a permanent dipole moment [20], where the DEP response of protein DEP is not relevant using the common DEP theory. As the conventional equation of DEP force heavily relies on the shape and size of the particles with induced dipole moment, the case is different with protein. This is because the aspect that could affect the DEP force includes the dielectric dispersions caused by the protein’s electrical double-layer, the permanent dipole characteristics, the hydration shell and the interaction of the protein with water, respectively.
The protein with the permanent dipole moment is the water molecule’s central orbit, as shown in Figure 2. This will be further discussed in the protein ‘Russian Doll’ theory proposed by Holzel and Pethig [21].

Figure 2. ‘Russian doll’ model of a protein. The protein is surrounded by a permanent dipole moment. The protein fills the most internal cavity which is strongly attached to water molecules.
The dipole field of protein projects outwards, surpassing the macroscopic boundary. According to Sayedi & Matyushov [22], proteins have a large amount of surface charge for their solubility in suspending medium. The negative and positive charges recompense to produce a net negative charge at their physiological state. However, the protein which is cytochrome-c studied by [22] is positively charged. The proteins’ irregular shape and non-balanced overall charge cause a huge dipole moment. The discussion on factors affecting DEP will be elaborated.
Several electrokinetics cases could highly affect DEP. As stated by [23], some of the factors are electroosmosis, thermal heat force (Joule Heating), electrokinetic force, hydrodynamic force, and Brownian motion. By minimizing the effect of these interferences, DEP force could be maximized. Electroosmosis is the fluid motion of the particles after being applied with frequency due to the viscosity of the medium [24]. The other case that could highly affect DEP is Joule Heating, also known as electrothermal motion, which is the movement in mediums due to the temperature gradient when electric current flows through an electrical conductor [25][26]. A study was conducted on the factors such as the diameter and distance of the electrode and channel height affecting Joule Heating [27].
Brownian motion is defined as the random movement of particles suspended in a medium [28]. This could also affect DEP response [29]. This phenomenon that needs to be overcome by DEP is the dispersive force linked to Brownian motion [20]. As the size of particles gets smaller to the nanometric scale, the DEP force becomes weaker, and it tends to be subjected to increasingly Brownian motion [30]. Thus, it is harder to manipulate using DEP as the size gets smaller.
The general phenomenon of Brownian motion in DEP is well-explained in the Fokker-Planck (or Smoluchowski) equation [31]. This equation is also known as the diffusion-advection equation. This equation has considered both pressure-driven and electrohydrodynamic flows for DEP simulation. The Brownian motion becomes weaker as it extends outwards the electrode region as the DEP force is strongest at the tip of the electrodes. DEP force can be neglected as it gets further from the electrodes, and the force of liquid diffusion takes over. The distribution of the Brownian motion compared with DEP force is visualized in Figure 3 below.
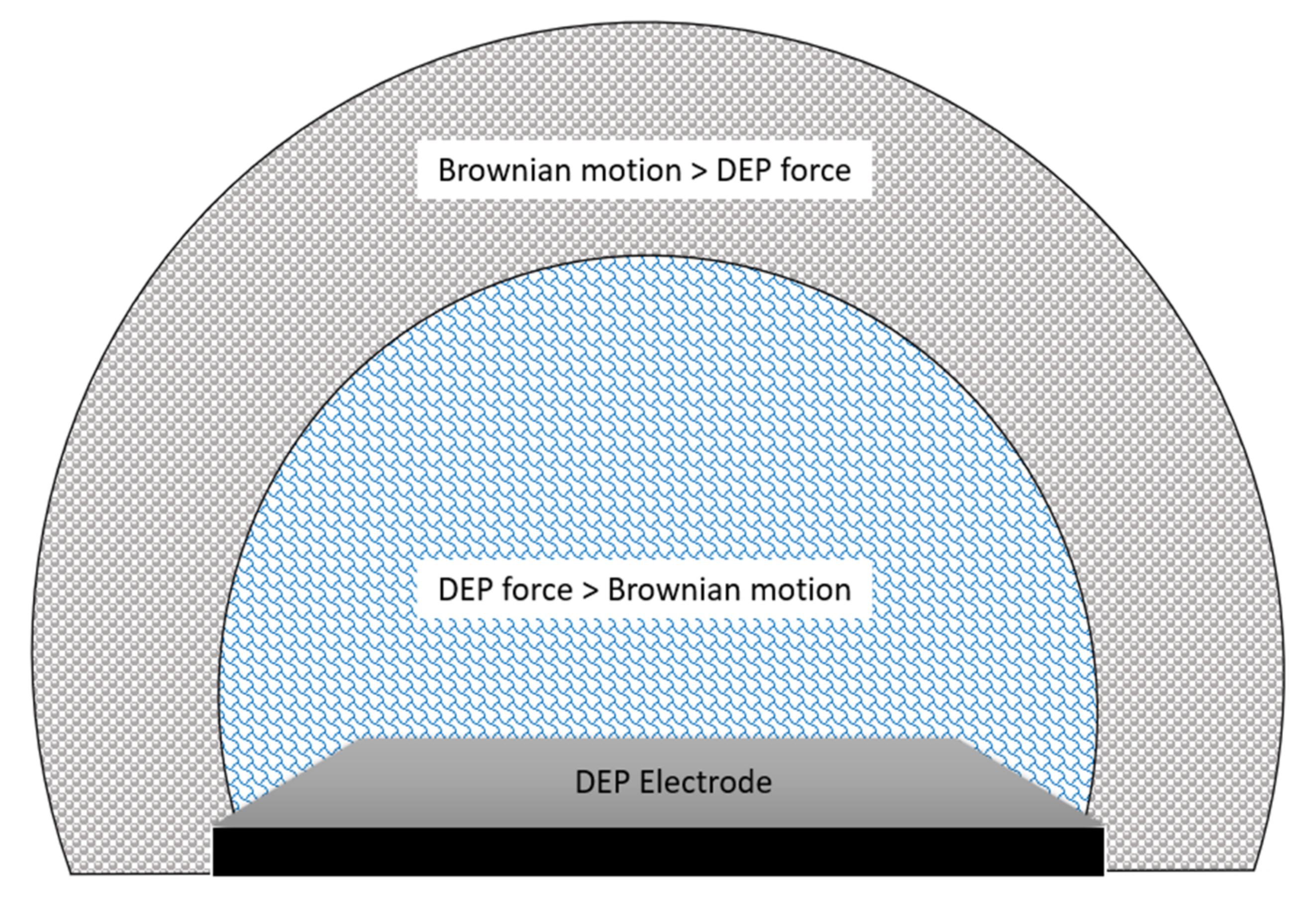
Figure 3. DEP force versus Brownian motion in terms of area of force concentration.
2. DEP Studies on Protein Albumin
Bovine serum albumin (BSA) is the most studied protein for DEP characterization. Basically, two types of electrodes configuration are being studied for BSA. The first type is insulator-based DEP (iDEP) and the other type is electrode-based DEP (eDEP). The definition of two types of DEP associated with studies with sample BSA is listed below in Table 1.
Table 1. Different types of DEP and their definition.
| Types of DEP | Definition | References |
|---|---|---|
| Insulator-based dielectrophoresis (iDEP) | An insulating layer or structured insulating arrangement is placed between the electrode where nonuniform fields are being created. Mostly applied with DC with high voltage. | [32][33][34][35] |
| Electrode-based dielectrophoresis (eDEP) | Nonuniform fields are created by metal-based, sharp-edged with narrow gaps spaced between the electrodes. | [36][37][38] |
The results obtained from albumin protein DEP studies and respected frequency are listed in Table 2 and Table 3 below:
Table 2. Insulator-based DEP type of electrode configuration studied for protein BSA.
| Frequency/ Type of Current |
DEP Principal Application | DEP Response |
Geometry of Electrodes | Conductivity of Suspending Medium |
Voltage and Frequency Applied | Reference |
|---|---|---|---|---|---|---|
| DC current | Aggregate trapping | pDEP | Elliptical post array | 50 µL phosphate buffer solution; 0.01 S/m | 1900 V/cm | [39] |
| DC current | Enrichment | pDEP | Circular array | Phosphate buffer solution; 0.01 S/m–0.04 S/m | 3000 V/cm | [40] |
| DC current | Enrichment | nDEP | Post array with ‘dove-tail’ geometry on the beginning of the post of both sides. | Deionized water; 25 to 100 S/cm | 700 V/cm | [41] |
| Both AC and DC current | Enrichment | nDEP | Parallel micro ridges | Phosphate buffered saline (PBS); 0.08–0.1 S/m | 5 V to 15 V; 10 Hz to 100 kHz | [42] |
Table 3. Electrode-based-DEP type of electrode configuration studied for protein BSA.
| Frequency/ Type of Current |
DEP Principal Application | DEP Response |
Geometry of Electrodes | Conductivity of Suspending Medium |
Voltage and Frequency Applied | Reference |
|---|---|---|---|---|---|---|
| AC current | Trapping | pDEP | Nanohole array | Water medium; 0.28 mS/m | 6 Vpp; 1 kHz | [43] |
| AC current | Enrichment | pDEP | Subarray | Ultrapure water; less than 1 S/cm | 0.7–14.1 Vrms; 10 kHz | [44] |
| AC current | Trapping | pDEP | Column-shaped nanopillar with a flat top | De-ionized water; 2 µS/cm | From 3 MV/m to 5 MV/m | [45] |
| AC current | Trapping | pDEP | Quadrupole | De-ionized water; 1 mS/m | Up to 8Vpp; 50 kHz and 5 MHz | [46] |
| AC current | Accumulation | pDEP | Nanocones | Water; 10−6 S/m | 10 V, 2.5 MHz | [47] |
Most of the studies of BSA were observed for pDEP but only two studies with nDEP of iDEP were reported [41][42]. According to Nakano et al. [39], nDEP results because the iDEP needs to overcome the problem of controlling the protein aggregates. Research has also been carried out to study the electrical properties of BSA by observing the impedance of BSA as BSA accumulates onto the electrodes [48].
The principle of eDEP is well-established as DEP was first found using the eDEP principle. iDEP was later developed through time [49]. As Benhal et al., (2020) stated, Joule heating and disintegration of electrodes are more likely to happen in eDEP as a higher gradient is being created. This could subsequently contaminate the biological sample [32]. According to Pethig, the usage of eDEP is more relevant for smaller molecules such as proteins, virions, vesicles and exosomes where high values of (E·∇)E are acquired, where E represents the electric field [37]. This is because the DEP manipulation of the targeted molecules will be based on the difference in dielectric properties of the membrane or ‘sheath’ and the charges of the inner cavity of the targeted particles.
Research has also been carried out by Washizu et al., (1994) on developing a protocol for identifying pDEP for protein for eDEP. The protocol describes that the first step is to identify the purity of the protein monomers using a gel chromatography method. Then, the concentration of the protein should be below 0.1 µg/mL. As for the conductivity of the suspending medium, the value should be below 1 mS/m [50]. This protocol has been widely used ever since for eDEP.
This entry is adapted from the peer-reviewed paper 10.3390/mi13081308
References
- Pohl, H.A. The motion and precipitation of suspensoids in divergent electric fields. J. Appl. Phys. 1951, 22, 869–871.
- Pohl, H.A. Some effects of nonuniform fields on dielectrics. J. Appl. Phys. 1958, 29, 1182–1188.
- Buyong, M.R.; Larki, F.; Takamura, Y.; Abd Aziz, N.; Yunas, J.; Hamzah, A.A.; Majlis, B.Y. Implementing the concept of dielectrophoresis in glomerular filtration of human kidneys. In Proceedings of the 2016 IEEE International Conference on Semiconductor Electronics (ICSE), Kuala Lumpur, Malaysia, 17–19 August 2016; pp. 33–37.
- Nasir, N.S.A.; Deivasigamani, R.; Rahim, M.K.A.; Nashruddin, S.N.A.M.; Hamzah, A.A.; Wee, M.F.M.R.; Buyong, M.R. Preliminary dielectrophoresis study: Manipulation of protein albumin and electrical quantification by using cyclic voltammetry technique. Microelectron. Int. 2021, 38, 162–171.
- Buyong, M.R.; Larki, F.; Faiz, M.S.; Hamzah, A.A.; Yunas, J.; Majlis, B.Y. A tapered aluminium microelectrode array for improvement of dielectrophoresis-based particle manipulation. Sensors 2015, 15, 10973–10990.
- Kua, C.; Lam, Y.C.; Yang, C.; Youcef-Toumi, K. Review of bio-particle manipulation using dielectrophoresis. Available online: https://dspace.mit.edu/handle/1721.1/7464 (accessed on 2 January 2021).
- Buyong, M.R.; Ismail, A.G.; Yunus, F.W.; Abd Samad, M.I.; Jamaludin, N.M.A.; Rahim, M.K.A.; Hamzah, A.A.; Majlis, B.Y.; Abd Aziz, N. Tapered dielectrophoresis microelectrodes: Device, operation, and application. In Proceedings of the 2019 IEEE Regional Symposium on Micro and Nanoelectronics (RSM), Putrajaya, Malaysia, 21–23 August 2019; pp. 172–175.
- Zhou, R.; Wang, P.; Chang, H.C. Bacteria capture, concentration and detection by alternating current dielectrophoresis and self-assembly of dispersed single-wall carbon nanotubes. Electrophoresis 2006, 27, 1376–1385.
- Buyong, M.R.; Larki, F.; Caille, C.E.; Abd Aziz, N.; Ismail, A.G.; Hamzah, A.A.; Majlis, B.Y. Dynamic dielectric properties characterization of tapered dielectrophoresis microelectrodes for selective detection and rapid manipulation of cells. Microelectron. Int. 2020, 37, 189–198.
- Qian, C.; Huang, H.; Chen, L.; Li, X.; Ge, Z.; Chen, T.; Yang, Z.; Sun, L. Dielectrophoresis for bioparticle manipulation. Int. J. Mol. Sci. 2014, 15, 18281–18309.
- Choi, W.; Min, Y.W.; Lee, K.Y.; Jun, S.; Lee, H.G. Dielectrophoresis-based microwire biosensor for rapid detection of Escherichia coli K-12 in ground beef. Lwt 2020, 132, 109230.
- Rashid, N.F.A.; Deivasigamani, R.; Wee, M.M.R.; Hamzah, A.A.; Buyong, M.R. Integration of a Dielectrophoretic Tapered Aluminum Microelectrode Array with a Flow Focusing Technique. Sensors 2021, 21, 4957.
- Rahim, M.K.A.; Jamaludin, N.M.A.; Santhanam, J.; Hamzah, A.A.; Buyong, M.R. Determination of Dielectrophoretic Unique Crossover Frequency by Velocity of Enterobacter Aerogenes Trajectory. Int. J. Nanoelectron. Mater. 2020, 13.
- Jamaludin, N.M.A.; Rahim, M.K.A.; Hamzah, A.A.; Abu, N.; Buyong, M.R. Optimization of the Isolation of Extracellular Vesicles via Dielectrophoresis: A preliminary Analysis. Int. J. Nanoelectron. Mater. 2020, 13, 129–142.
- Modarres, P.; Tabrizian, M. Alternating current dielectrophoresis of biomacromolecules: The interplay of electrokinetic effects. Sens. Actuators B Chem. 2017, 252, 391–408.
- Cottet, J.; Fabregue, O.; Berger, C.; Buret, F.; Renaud, P.; Frénéa-Robin, M. MyDEP: A new computational tool for dielectric modeling of particles and cells. Biophys. J. 2019, 116, 12–18.
- Honda, C.; Kamizono, H.; Samejima, T.; ENDO, K. Studies on thermal aggregation of bovine serum albumin as a drug carrier. Chem. Pharm. Bull. 2000, 48, 464–466.
- Yang, C.; Lei, U. Quasistatic force and torque on ellipsoidal particles under generalized dielectrophoresis. J. Appl. Phys. 2007, 102, 094702.
- Pethig, R. Limitations of the Clausius-Mossotti function used in dielectrophoresis and electrical impedance studies of biomacromolecules. Electrophoresis 2019, 40, 2575–2583.
- Pethig, R. Protein Dielectrophoresis: A Tale of Two Clausius-Mossottis—Or Something Else? Micromachines 2022, 13, 261.
- Hölzel, R.; Pethig, R. Protein dielectrophoresis: I. Status of experiments and an empirical theory. Micromachines 2020, 11, 533.
- Seyedi, S.S.; Matyushov, D.V. Protein dielectrophoresis in solution. J. Phys. Chem. B 2018, 122, 9119–9127.
- Nakano, A.; Ros, A. Protein dielectrophoresis: Advances, challenges, and applications. Electrophoresis 2013, 34, 1085–1096.
- Nakano, A. Protein Dielectrophoresis Using Insulator-Based Microfluidic Platforms; Arizona State University: Tempe, AZ, USA, 2014.
- Yan, Y.; Guo, D.; Wen, S. Joule heating effects on two-phase flows in dielectrophoresis microchips. BioChip J. 2017, 11, 196–205.
- Abd Rahman, N.; Ibrahim, F.; Yafouz, B. Dielectrophoresis for biomedical sciences applications: A review. Sensors 2017, 17, 449.
- Wang, Y.; Du, F.; Baune, M.; Thöming, J. Predicting and eliminating Joule heating constraints in large dielectrophoretic IDE separators. Chem. Eng. Sci. 2015, 137, 235–242.
- Zaman, M.A.; Wu, M.; Padhy, P.; Jensen, M.A.; Hesselink, L.; Davis, R.W. Modeling brownian microparticle trajectories in lab-on-a-chip devices with time varying dielectrophoretic or optical forces. Micromachines 2021, 12, 1265.
- Kadaksham, A.T.; Singh, P.; Aubry, N. Dielectrophoresis of nanoparticles. Electrophoresis 2004, 25, 3625–3632.
- Altintas, E.; Böhringer, K.F.; Fujita, H. Micromachined Linear Brownian Motor: Transportation of Nanobeads by Brownian Motion Using Three-Phase Dielectrophoretic Ratchet. Jpn. J. Appl. Phys. 2008, 47, 8673.
- Midelet, C.; Le Pioufle, B.; Werts, M. Brownian Motion and Large Electric Polarizabilities Facilitate Dielectrophoretic Capture of Sub-200 nm Gold Nanoparticles in Water. ChemPhysChem 2019, 20, 3354–3365.
- Benhal, P.; Quashie, D.; Kim, Y.; Ali, J. Insulator Based Dielectrophoresis: Micro, Nano, and Molecular Scale Biological Applications. Sensors 2020, 20, 5095.
- Gallo-Villanueva, R.C.; Sano, M.B.; Lapizco-Encinas, B.H.; Davalos, R.V. Joule heating effects on particle immobilization in insulator-based dielectrophoretic devices. Electrophoresis 2014, 35, 352–361.
- Nakidde, D.; Zellner, P.; Alemi, M.M.; Shake, T.; Hosseini, Y.; Riquelme, M.V.; Pruden, A.; Agah, M. Three dimensional passivated-electrode insulator-based dielectrophoresis. Biomicrofluidics 2015, 9, 014125.
- Jen, C.-P.; Chen, T.-W. Selective trapping of live and dead mammalian cells using insulator-based dielectrophoresis within open-top microstructures. Biomed. Microdevices 2009, 11, 597–607.
- Zellner, P.; Shake, T.; Sahari, A.; Behkam, B.; Agah, M. Off-chip passivated-electrode, insulator-based dielectrophoresis (OπDEP). Anal. Bioanal. Chem. 2013, 405, 6657–6666.
- Pethig, R. Where is dielectrophoresis (DEP) going? J. Electrochem. Soc. 2016, 164, B3049.
- Shayestehpour, H.; Nassiri Nazif, K.; Soufi, A.; Saidi, M. Proposing a high-efficiency dielectrophoretic system for separation of dead and live cells. Sci. Iran. 2018, 25, 186–195.
- Nakano, A.; Chao, T.C.; Camacho-Alanis, F.; Ros, A. Immunoglobulin G and bovine serum albumin streaming dielectrophoresis in a microfluidic device. Electrophoresis 2011, 32, 2314–2322.
- Nakano, A.; Camacho-Alanis, F.; Chao, T.; Ros, A. Systematic investigation of insulator-based protein dielectrophoresis under DC condition. In Proceedings of the 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences 2011, MicroTAS 2011, Seattle, WA, USA, 2–6 October 2011; pp. 644–646.
- Lapizco-Encinas, B.H.; Ozuna-Chacón, S.; Rito-Palomares, M. Protein manipulation with insulator-based dielectrophoresis and direct current electric fields. J. Chromatogr. A 2008, 1206, 45–51.
- Zhang, P.; Liu, Y. DC biased low-frequency insulating constriction dielectrophoresis for protein biomolecules concentration. Biofabrication 2017, 9, 045003.
- Barik, A.; Otto, L.M.; Yoo, D.; Jose, J.; Johnson, T.W.; Oh, S.-H. Dielectrophoresis-enhanced plasmonic sensing with gold nanohole arrays. Nano Lett. 2014, 14, 2006–2012.
- Laux, E.M.; Knigge, X.; Bier, F.F.; Wenger, C.; Hölzel, R. Dielectrophoretic immobilization of proteins: Quantification by atomic force microscopy. Electrophoresis 2015, 36, 2094–2101.
- Yamamoto, T.; Fujii, T. Active immobilization of biomolecules on a hybrid three-dimensional nanoelectrode by dielectrophoresis for single-biomolecule study. Nanotechnology 2007, 18, 495503.
- Zheng, L.; Brody, J.P.; Burke, P.J. Electronic manipulation of DNA, proteins, and nanoparticles for potential circuit assembly. Biosens. Bioelectron. 2004, 20, 606–619.
- Schäfer, C.; Kern, D.P.; Fleischer, M. Capturing molecules with plasmonic nanotips in microfluidic channels by dielectrophoresis. Lab A Chip 2015, 15, 1066–1071.
- Mohamad, A.S.; Hamzah, R.; Hoettges, K.F.; Hughes, M.P. A dielectrophoresis-impedance method for protein detection and analysis. AIP Adv. 2017, 7, 015202.
- Hill, N.; Lapizco-Encinas, B.H. Continuous flow separation of particles with insulator-based dielectrophoresis chromatography. Anal. Bioanal. Chem. 2020, 412, 3891–3902.
- Washizu, M.; Suzuki, S.; Kurosawa, O.; Nishizaka, T.; Shinohara, T. Molecular dielectrophoresis of biopolymers. IEEE Trans. Ind. Appl. 1994, 30, 835–843.
This entry is offline, you can click here to edit this entry!
