Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Genetics & Heredity
RNA fragments derived from full-length small nucleolar RNAs (snoRNAs) have been shown to be specifically excised and functional. These sno-derived RNAs (sdRNAs) have been implicated as gene regulators in a multitude of cancers, controlling a variety of genes post-transcriptionally via association with the RNA-induced silencing complex (RISC).
- cancer
- gene regulation
- small nucleolar RNA (snoRNA)
- small nucleolar derived RNA (sdRNA)
- microRNA (miRNA)
- RNA
- genetics
1. snoRNA Structure and Function
One class of ncRNA found to give rise to functional ndRNAs is the snoRNA. SnoRNAs constitute a class of ncRNA localized to the nucleolus that can be further divided into two main subgroups: C/D box and H/ACA box snoRNA [3]. C/D box snoRNA are structurally distinguished by the presence of a 5′ box C motif (RUGAUGA, R = purine) and its C’ box partner, as well as a 3′ box D motif (CUGA) with its D’ box partner. Self-complementarity drives the formation of the C/D box snoRNA stem structure that in turn binds accessory proteins NOP56, NOP58, 15.5 K, and fibrillarin to form the functional sno-ribonucleoprotein (snoRNP) (Figure 1 and Figure 2). Situated in the nucleolus, antisense domains located within the C/D box snoRNA serve as guides for the snoRNP to bind target ribosomal RNAs (rRNAs). The methyltransferase fibrillarin is then responsible for carrying out 2′-O-ribose-methylation of the rRNA [4,5,6]. H/ACA box snoRNA contain a 3′ ACA box (ACANNN, N = any nt) and two hairpins linked by an H box (ANANNA, N = any nt). The H/ACA box snoRNP complex is formed by association with NHP2, NOP10, GAR1, and dyskerin proteins (Figure 1 and Figure 3). The H/ACA snoRNA hairpins guide the snoRNP to target rRNAs where dyskerin carries out pseudouridylation [7,8,9]. Erroneous expression and/or activity of either of these classes of snoRNA have been extensively linked to tumorigenesis. However, reports have implied that several proteins comprising the snoRNP are required for the biogenesis of distinct species of sdRNA [10].
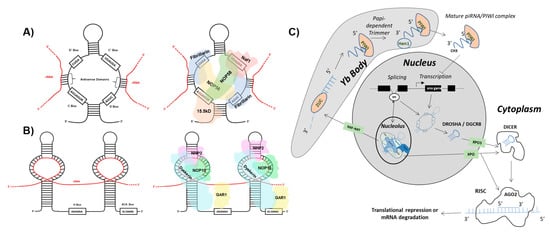
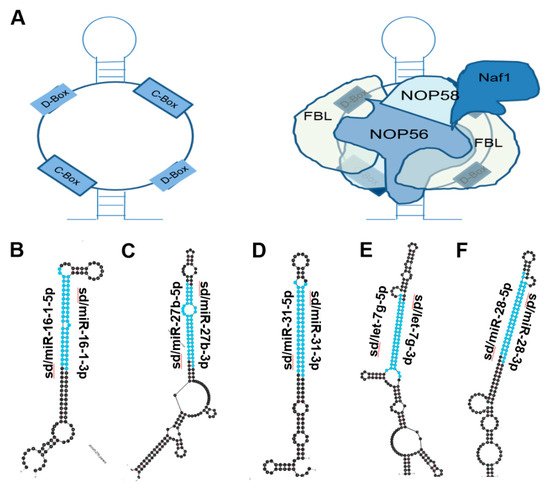
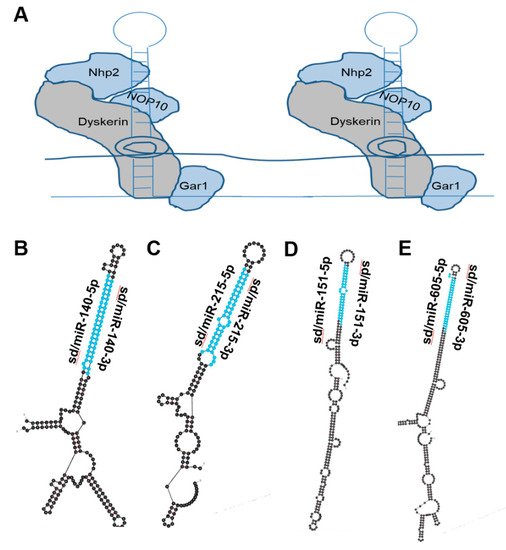

Figure 1. sdRNAs arise from full-length snoRNAs. (A) C/D box snoRNA structure and accessory proteins. (Left) The C/D box snoRNA consists of a 5′ C box (RUGAUGA) motif, a 3′ D box (CUGA) motif, and the C’ and D’ boxes located internally. Antisense domains identify target rRNAs (red) via complementarity. (Right) The C/D box snoRNP complex consists of NOP56, NOP58, 15.5 K, and fibrillarin proteins [4,5,6]. (B) H/ACA box snoRNA structure and accessory proteins. (Left) The H/ACA box snoRNA consists of a 3′ ACA box (ACANNN, N = any nt) and two hairpins that target rRNA (red) linked by an H box (ANANNA, N = any nt). (Right) The H/ACA box snoRNP complex consists of NHP2, NOP10, GAR1, and dyskerin proteins [7,8,9]. (C) sdRNA biogenesis and function. Biogenesis of Argonaute 2 (AGO2)-associating sno-derived RNAs (sdRNAs) Full length small-nucleolar RNAs (snoRNAs) are generated either as products of transcription or splicing [11,12,13,14]. snoRNAs produced by transcription can give rise to microRNA-like sdRNAs which are specifically excised from parent snoRNA transcripts by employment of the classical microRNA (miRNA) processing pathway. This occurs by processing of parent snoRNAs into smaller transcripts by the microprocessor complex which consists of Drosha Ribonuclease III (DROSHA) and DiGeorge syndrome critical region 8 (DGCR8). The intermediate snoRNA then undergoes cytoplasmic exportation via exportin 5 (XPO5). Following this, the smaller cytoplasmic snoRNA is processed by Dicer RNase III endonuclease (DICER) to generate the mature sdRNA which associates with AGO2, leading to the formation of the RNA-induced Silencing Complex (RISC). Similar to miRNAs, these sdRNAs function in post-transcriptional gene suppression by antisense binding to target mRNA transcripts within RISC [15]. That said, snoRNAs produced by splicing can also enter the classical miRNA processing pathway. Spliced snoRNAs, however, can bypass processing from DROSHA/DGCR8 and/or DICER as a result of trafficking to the nucleolus and subsequent processing by the fibrillarin complex followed by cytoplasmic export via a transporter belonging to the Exportin (XPO) family of proteins [10]. Biogenesis of PIWI-associated RNA (piRNA) like sdRNAs Spliced snoRNAs which arrive at the nucleolus for fibrillarin processing can be trafficked into Yb bodies via Nuclear RNA Export Factor (NXF1)/ Nuclear Transport Factor 2 Like Export Factor 1 (NXT1) where the 3′ end is cleaved by Zucchini (ZUC) and subsequently degraded. The remaining transcript is processed further within the Yb body by the papi-dependent trimmer. Following this, HEN1 double-stranded RNA binding protein binds at the 3′ end of the transcript where it adds a methyl group, generating a mature piRNA/PIWI complex which is exported to the cytoplasm. This piRNA/PIWI complex can then be shuttled back into the nucleus where it functions to inhibit transcription [16,17].

Figure 2. miRNAs derived from C/D Box snoRNA-like transcripts that bind fibrillarin. (A) Cartoon illustration of C/D Box snoRNA (left) and schematic of C/D Box snoRNA bound to fibrillarin complex, giving rise to snoRNP (right). Fibrillarin complex consists of NOP56, NOP58, NAF1 and Fibrillarin [18]. (B–F) Most thermodynamically stable secondary structures of C/D Box snoRNA-like transcripts described to bind fibrillarin complex [19] were obtained from mfold [20]. Mature miRNA sequences embedded in these transcripts are highlighted in blue. (B) miR16-1 (ENSG00000208006) is downregulated in chronic lymphocytic leukemia, gastric, NSCLC, Osteosarcoma, and breast cancer and is embedded within a C/D Box snoRNA-like transcript (GRCh38: chr 13:50048958-50049077:-1) known to bind fibrillarin [1,21,22,23,24]. (C) miR-27b (ENSG00000207864) is downregulated in prostate, lung, and bladder cancer and is located within a C/D Box snoRNA-like transcript (GRCh38: chr 9:95085436-95085592:1) known to bind fibrillarin [25,26,27]. (D) miR-31 (ENSG00000199177) is upregulated in colorectal, HNSCC, and lung cancer but is downregulated in glioblastoma, melanoma, and prostate cancer. miR31 is also located within a C/D Box snoRNA-like transcript (GRCh38: chr 9:21512102-21512221:-1) known to bind fibrillarin [28,29,30,31,32,33]. (E) miR-let7g (ENSG00000199150) has been shown to be downregulated in NSCLC, colorectal and ovarian cancers and is also embedded within a C/D Box snoRNA-like transcript (GRCh38: chr 3:52268239-52268408:-1) known to bind fibrillarin [34,35,36]. (F) miR-28 (ENSG00000207651) has been shown to be downregulated in B-cell lymphoma, prostate and breast cancer and is located within a C/D Box snoRNA-like transcript (GRCh38: chr 3:188688746-188688887:1) known to bind fibrillarin [37,38,39].

Figure 3. miRNAs derived from Box H/ACA snoRNA-like transcripts that bind dyskerin. (A) Schematic of box H/ACA snoRNA bound to dyskerin complex, giving rise to snoRNP. Dyskerin complex includes Dyskerin, NHP2, NOP10, and GAR1 [18]. (B–E) Most thermodynamically stable secondary structures of box H/ACA snoRNA-like transcripts known to bind dyskerin complex [40] were generated by mfold [20]. Mature miRNA sequences derived from these transcripts are highlighted in blue. (B) Box H/ACA snoRNA-like transcript (GRCh38: chr 16:69933072-69933264:1) was identified to bind dyskerin and encompasses miR-140 (ENSG00000208017) which functions as a tumor-suppressing RNA and is downregulated in prostate cancer [41]. (C) Box H/ACA snoRNA-like transcript (GRCh38: chr1:220117845-220118007:-1) was identified to bind dyskerin and surrounds miR-215 (ENSG00000207590) which is known to function as a tumor suppressor and is downregulated in ovarian, colorectal, prostate and lung cancer [42,43]. (D) Box H/ACA snoRNA-like transcript (GRCh38: chr8:140732552-140732807:-1) was determined to bind dyskerin and encapsulates miR-151 (ENSG00000254324) which is recognized as a tumor-suppressing RNA and is downregulated in prostate cancer [44,45,46]. (E) Box H/ACA snoRNA-like transcript (GRCh38: chr 10:51299393-51299708:1) was determined to bind dyskerin and surrounds miR-605 (ENSG00000207813) which is described to act as a tumor-suppressing RNA in melanoma, colorectal, breast, lung and prostate cancer [47,48,49,50,51].
2. sdRNAs in Cancer
2.1. sd/miR-664
In 2019 Lv et al. published a paper focused on elucidating the tumor-suppressive role of sd/miR-664 in cervical cancer. Comparing sd/miR-664 expression in cervical cancer patient samples and control tissue revealed that the sdRNA is expressed at lower levels in cancer. Using the Si-Ha cell line as a model of cervical cancer, sd/miR-664 expression was found to enhance apoptosis and reduce cell viability and migration. In a Si-Ha mouse xenograft model, treatment with an sd/miR-664 mimic drastically reduced tumor volume and weight while enhancing tumor apoptosis. The binding target was predicted to be c-Kit using online target prediction software, and this was confirmed via the Renilla luciferase assay. Sd/miR-664 expression was negatively correlated with c-Kit expression, which further solidified the proto-oncogene c-Kit as a target of this tumor-suppressive sdRNA in cervical cancer [57].
To assess the role of sd/miR-664 in cutaneous squamous cell carcinoma (cSCC), Li et al. quantified the sdRNA’s expression by in situ hybridization in patient samples as well as cSCC cell lines [58]. Both analyses revealed an increase in sd/miR-664 in cSCC compared to control. Phenotypic assays showed increased cSCC viability, colony formation, invasion, and migration when sd/miR-664 levels were elevated in cell lines. Additionally, a murine cSCC xenograft model treated with sd/miR-664 mimic exhibited increased tumor volume compared to control. The 3′-UTR of IRF2 was predicted and then confirmed to be a binding target of sd/miR-664. Taken together, sd/miR-664 functions as an onco-miR in cSCC by downregulating IRF2 expression. While the role of IRF2 has not been established in cSCC, IRF2 has been identified as a tumor suppressor in lung cancer and gastric cancer and, paradoxically, as an oncogene in testicular embryonal carcinoma [78,79,80]. This adds a layer of complexity to sd/miR-664, showing that its overall impact on tumor progression can be tissue context-dependent.
2.2. Sd/miR-1291
Building on their 2016 publication where they found sd/miR-1291 to be downregulated in pancreatic cancer patient tissues, Tu et al. published an article in 2020 to further investigate this mechanism of action in pancreatic cancer cells [59,60]. They found that increasing sd/miR-1291 reduced ASS1 levels in the ASS1-abundant L3.3 pancreatic cancer cell line and sensitized them to arginine deprivation in vitro. In addition, the glucose transporter GLUT1 mRNA has previously been shown to be a direct target of sd/miR-1291 in renal cell carcinoma (RCC), and Tu et al. confirmed this to be the case in pancreatic cancer cells as well with a resulting decrease in glycolytic capacity [61]. Further, sd/miR-1291 treatment sensitized pancreatic cancer cell lines to cisplatin. Taken together, sd/miR-1291 has a clear role as a modulator of cell metabolism to bring about tumor suppression in pancreatic cancer.
2.3. Sd/miR-3651
A 2020 publication aimed at identifying novel contributors to colorectal cancer (CRC) found that miR-3651, which arises from SNORA84, was overexpressed in 34/40 CRC patient tumors [65]. Model cell lines of CRC were observed to have ~3-fold overexpression of sd/miR-3651. Phenotypic assays revealed that sd/miR-3651 expression was positively correlated with CRC cell proliferation, and that reduction of the sdRNA enhanced apoptosis via deactivation of PI3K/AKT and MAPK/ERK signaling. Target prediction identified the 3′-UTR of tumor suppressor TBX1 as a sd/miR-3651 target, which was confirmed via the luciferase assay. Since TBX1 is a transcription factor that has an established role as a regulator of PI3K/AKT and MAPK/ERK pathways, these results show a coherent mechanism of action by which sd/miR-3651 inactivates TBX1 and thereby activates pro-growth mitogenic pathways to function as an oncogenic sdRNA.
2.4. sd/miR-768
In 2012, Su et al. published a study in which they interrogated gastric cancer patient samples for miRNA expression. They found that sd/miR-768-5p, which arises from SNORD71 (Figure 4), was significantly downregulated in cancer samples [68]. A 2013 study by Blenkiron et al. identified sd/miR-768-5p as a YB-1 binding partner via CO-IP in MCF7 cells [67]. The authors proposed potential explanations for the YB-1/sdRNA interaction including YB-1 functioning as a ncRNA-sponge to prevent ncRNA-mediated transcriptional repression as well as YB-1 potentially playing a role in ncRNA biogenesis.

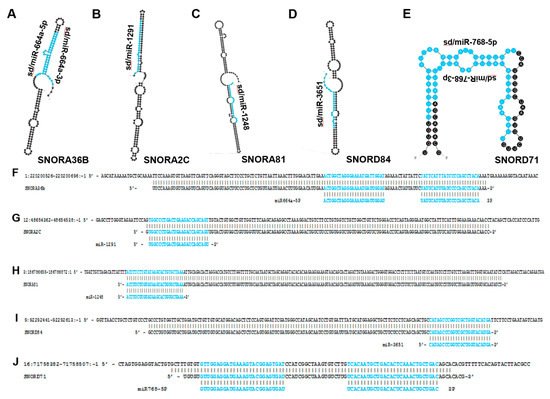
Figure 4. miRNAs that arise from snoRNA transcripts. The most thermodynamically stable secondary structures of snoRNA transcripts were generated by mfold [20]. Annotated miRNA sequences embedded in these transcripts are highlighted in blue (A–E). (A) miR-664a is embedded within the SNORA36B transcript and has been shown to function as a tumor-suppressing RNA in HCC and some types of cervical cancer while also functioning as a tumor-promoting RNA in cervical squamous cell carcinoma [56]. (B) miR-1291 is located within the SNORA2C transcript and is characterized as a tumor-suppressing RNA in pancreatic, prostate, breast and renal cell carcinoma [59,60,61,62,63]. (C) miR-1248 is derived from SNORA81 and has been determined to be upregulated in prostate cancer [64]. (D) miR-3651 is embedded within SNORA84 and is upregulated in colorectal cancer but downregulated in esophageal cancer [65,66]. (E) miR-768 resides within the SNORD71 transcript and has been shown to be downregulated in lung, breast, ovary, liver, parotid gland, thyroid gland cancers and melanoma [67,68,69]. (F–J) Mature miRNA sequences (highlighted in blue) were aligned to their corresponding snoRNA precursor transcripts and snoRNAs were aligned to their respective genomic regions. (F) Alignment between human genome (GRCh38: chr1:220200526-220200696:-1) (top), SNORA36B (ENSG00000222370) (middle), and miR664a (ENSG00000281696) (bottom). (G) Alignment between human genome (GRCh38: chr12:48654362-48654538:-1) (top), SNORA2C (ENSG00000221491) (middle) and miR-1291 (ENSG00000281842) (bottom). (H) Alignment between human genome (GRCh38: chr 3:186786655-186786872:1) (top), SNORA81 (ENSG00000221420) (middle), and miR-1248 (ENSG00000283958) (bottom). (I) Alignment between human genome (GRCh38: chr 9:92292441-92292613:-1) (top) SNORA84 (ENSG00000239183) (middle) and miR-3651 (ENSG00000281156) (bottom). (J) Alignment between human genome (GRCh38: chr 16:71758382-71758507:-1) (top), SNORD71 (ENSG00000223224) (middle) and miR-768 (ENSG00000223224) (bottom).
2.5. sd/miR-1248
There is a clear need for additional reliable biomarkers to aid care providers in their decision on which treatment to apply to prostate cancer patients following radical prostatectomy. Aimed at meeting this need, Pudova et al. published a paper in 2020 focused on identifying differentially expressed miRNAs between prostate cancer patient tumors with or without lymphatic dissemination [64]. Among the nine miRNAs found to be significantly upregulated in prostate cancer with lymphatic dissemination, sd/miR-1248 was identified which arises completely from SNORA81.
2.6. hsa-sno-HBII-296B and hsa-sno-HBII-85-29
In 2015, Müller et al. performed small RNA sequencing on six PDAC tissue samples and five normal pancreas tissue samples to assess the expression of ncRNAs not typically found via microarray analysis [70]. They considered 45 noncoding RNAs identified as significantly down-regulated in PDAC, of which there were fourteen sdRNAs and a single sno-derived piRNA. The most downregulated sdRNA in PDAC, as determined by log2 fold change, was hsa-sno-HBII-85-29. Four sdRNAs of a total 78 ncRNAs were significantly upregulated in PDAC. The most upregulated sdRNA in PDAC, as determined by log2 fold change, was hsa-sno-HBII-296B. Subsequent analyses were focused on a miRNA identified in the study, however this work constitutes the only publication that characterizes differentially expressed sdRNAs focused specifically in PDAC. PDAC is a particularly difficult cancer to detect and treat, with a reported 5-year survival rate of just 11% [82].
4.7. Sno-miR-28
By measuring snoRNA expression following induced P53 activation, Yu et al. found significant downregulation of all snoRNAs associated with the SNGH1 snoRNA gene [71]. One sdRNA arising from SNGH1-associated sno28, sno-miR-28, was identified in complex with AGO via HITS-CLIP data and was found to be abundantly expressed in patient breast cancer tissue. Elevating TP53 expression in vitro consequently reduced sno-mir-28, and in silico and in vitro target engagement experiments revealed the P53-stabilizing protein TAF9B as a likely sno-mir28 target. Taken together, the authors proposed that sno-mir28 directly regulates TAF9B to bring about indirect repression of P53, forming a loop as P53 overexpression decreases sno-mir28 levels. Taqman expression analysis of matched breast cancer patient tumor and normal tissues revealed that SNHG1, SNORD28 and sno-miR-28 were all significantly upregulated in tumors. Furthermore, using the MCF10A cell line as a model of undifferentiated breast epithelium, the authors found that sno-miR-28 overexpression enhanced breast cancer cells’ proliferative capacity and colony formation. In sum, this study defined a role for sno-miR-28 as an oncogenic sdRNA heavily involved in suppressing the P53 pathway.
2.8. sdRNA-93
In 2017 our lab published a study in which we investigated the role of sdRNAs in the aggressive breast cancer phenotype [72]. RNAseq analysis identified 13 total full length snoRNAs differentially expressed (>7.5×) in MDA-MB-231 compared to MCF7, 10 of which consistently give rise to sdRNAs that associate with AGO in publicly available HITS-CLIP data. We elected to focus on the sdRNA arising from sno93 (sdRNA-93) due to it displaying the highest differential expression in MDA-MB-231 (≥75×) and previous publications implicating sdRNA-93 with miRNA-like silencing capabilities in luciferase assays [83]. SdRNA-93 silencing in MDA-MB-231 reduced cell invasion by >90% while sdRNA-93 overexpression conversely enhanced cell invasion by >100% at the same time point. By contrast, sdRNA-93 silencing in MCF7 had no significant effect on invasion while overexpression resulted in a substantial ~80% increase in cell invasion. We then employed multiple in silico miRNA target-prediction algorithms which predicted with consensus that Pipox, a gene involved in sarcosine metabolism which has been implicated in breast cancer progression, is a likely candidate for regulation by sdRNA-93.
2.9. sdRNA-D19b and sdRNA-A24
A study published in 2022 by our lab identified 38 specifically-excised and differentially-expressed sdRNAs in prostate cancer [73]. We began by querying PCa patient TCGA datasets alongside TCGA-normal prostate using an in-house web-based search algorithm SURFR (Short Uncharacterized RNA Fragment Recognition). SURFR aligns next generation sequencing (NGS) datasets to a frequently updated database of all human ncRNAs, performs a wavelet analysis to specifically determine the location and expression of ncRNA-derived fragments (ndRNAs) and then conducts an expression analysis to identify significantly differentially expressed ndRNAs. Two sdRNAs, sdRNA-D19b and sdRNA-A24 (Figure 3), were among the most overexpressed in PCa patient tumors and were identified as AGO-associated in publicly available datasets so they were selected for further scrutiny. In vitro phenotypic assays in PC3 cells, a model cell line for castration-resistant prostate cancer (CRPC), revealed that both sdRNAs markedly increased PC3 cell proliferation and that sdRNA-D19b, in particular, greatly enhanced cell migration. When the sdRNAs were overexpressed alongside treatment with chemotherapeutic agents, both sdRNAs provided drug-specific resistances with sdRNA-D19b levels correlating with paclitaxel resistance and sdRNA-A24 conferring dasatinib resistance. Multiple in silico target-prediction algorithms provided a consensus prediction of the CD44 and CDK12 3′ UTRs as targets for sdRNA-D19b and sdRNA-A24 respectively, which we then confirmed in vitro via the Renilla luciferase assay. Taken together, this work outlines a biologically coherent mechanism by which sdRNAs downregulate tumor suppressors in CRPC to enhance proliferative/metastatic capabilities and to encourage chemotherapeutic resistance.
2.10. Sd/miR-140
Based on prior investigations linking high expression of the long-noncoding RNA MALAT1 with poor prostate cancer patient prognosis, Hao et al.’s 2020 paper focused on elucidating the mechanism of action by which MALAT1 brings about this effect [41]. In silico target prediction and in vitro confirmation via the Renilla luciferase assay identified miR-140 as a target of MALAT1, indicating that MALAT1 functions as a miRNA sponge to reduce sd/miR-140 bioavailability. This was further supported by RIP-seq analysis where direct engagement of MALAT1 and sd/miR-140 was confirmed. Target prediction indicated that sd/miR-140’s target is the 3′UTR of BIRC, and this was confirmed via the Renilla luciferase assay. Knockdown of MALAT1 inhibited the growth of prostate cancer both in vitro and in vivo, an effect that the authors suggest is brought about through sd/miR-140’s release from repression by MALAT1. This outlines a pathway where sd/miR-140 functions as a tumor-suppressing RNA and is tightly regulated by the lncRNA MALAT1.
2.11. Sd/miR-151
A study in 2020 by Chen et al. found that sd/miR-151 is downregulated in human prostate cancer cell lines [44]. In prostate cancer cells, Chen et al. demonstrated that sd/miR-151 overexpression inhibited cell proliferation, migration, and invasion; enhanced apoptosis; and sensitized cells to treatment with 5-FU, an antimetabolite chemotherapy. While no target prediction was performed, the authors did find that overexpression of sd/miR-151 reduces phosphorylation of PI3K/AKT. Even without a precisely mapped mechanism of action, this publication links sd/miR-151 with prostate cancer as a tumor-suppressive sdRNA.
2.12. Sd/miR-215
A 2015 study by Ge et al. found that sd/miR-215 expression was reduced in both epithelial ovarian cancer cell lines and patient tissue [42]. Sd/miR-215 decreased cell proliferation, enhanced apoptosis, and enhanced sensitivity to paclitaxel treatment. In addition, increased sd/miR-205 resulted in a decrease of cellular X-linked inhibitor of apoptosis (XIAP) mRNA albeit without confirmation of target engagement. Vychytilova-Faltejskova et al. found in 2017 that sd/miR-215-5p is significantly reduced in CRC patient samples and that low sd/miR-215-5p expression is associated with late clinical stages of CRC as well as poor overall survival for CRC patients [43]. Overexpression of sd/miR-215-5p reduced cell proliferation, viability, colony formation, invasion, migration, in vivo tumor volume, and weight while enhancing apoptosis. Epiregulin (EREG) and HOXB9 mRNA were confirmed to be two targets of sd/miR-215-5p. Both genes are involved in epithelial growth factor receptor (EGFR) signaling, a critical pathway exploited by CRC to promote tumor growth.
2.13. Sd/miR-605
With their 2011 publication, Xiao et al. found that sd/mir-605 overexpression significantly reduced cell survival and enhanced apoptosis in breast and colon cancer cell lines [47]. The MDM2 mRNA was predicted as a target of sd/mir-605, and sd/mir-605 overexpression was found to cause a reduction in MDM2 protein. MDM2 is a regulator of P53 tumor suppressor, and sd/mir-605 transfection was shown to increase P53 activity without increasing P53 protein levels, suggesting the sdRNA relieves MDM2-mediated repression of P53. In 2014 a study by Huang et al. found that single nucleotide polymorphisms (SNPs) in the sd/miR-605 precursor were significantly correlated with a shorter prostate cancer biochemical recurrence for patients [48].
2.14. Sd/miR-16-1
S/dmiR-16-1 is one of the miRNAs first implicated in human cancer [1]. In 2002, a paper by Calin et al. found that the sd/miR-16-1 gene was deleted in more than 65% of chronic lymphocytotic leukemia (CLL). Northern blot analysis of sd/miR-16-1 expression in CLL patient cancer and normal tissue revealed that miR-16-1 is downregulated in cancer. Sd/miR-16-1 has since been verified as a tumor suppressor in a multitude of cancers, several of which have been detailed in this 2009 review by Aqeilan et al. [84]. In 2017, the sdRNA was demonstrated to function as a tumor-suppressing RNA in gastric cancer by Wang et al. [21]. The authors found sd/miR-16-1 to be downregulated in gastric cancer patient samples. Additionally, overexpression decreased gastric cancer cell migration, invasion, and colony formation in vitro while reducing tumor volume in vivo. Target engagement with the 3′-UTR of TWIST1 was confirmed via the Renilla luciferase assay. Interestingly, a 2018 paper by a separate group, Feng et al., discovered that sd/miR-16-1 also targets TWIST1 in non-small cell lung cancer (NSCLC) [22]. Overexpression inhibited NSCLC proliferation, migration, and invasion in vitro. Together these publications add to the numerous examples of sd/miR-16-1 functioning as a tumor-suppressing RNA.
2.15. Sd/miR-27b
In 2012, Ishteiwy et al. found that sd/miR-27b is downregulated in castration-resistant prostate tumors compared to primary prostate cancer and normal tissue [25]. Increasing sd/miR-27b expression in CRPC cell lines reduced invasion, metastasis, and colony formation. While there was no confirmation of target interactions, the authors found that modulating sd/miR-27b expression had a subsequent impact on Rac1 activity and E-cadherin expression. These findings indicate a tumor-suppressive role for sd/miR-27b in CRPC whose mechanism may involve the tumor suppressor gene E-cadherin.
2.16. Sd/miR-31
The diverse role of miR-31 in cancer has been well studied and is the subject of two excellent reviews both in 2013 and more recently in 2018 [28,29]. Some examples from the aforementioned reviews include sd/miR-31 acting as a tumor-promoting RNA in CRC, HNSCC, and lung cancer while acting as a tumor-suppressing RNA in glioblastoma, melanoma, and prostate cancer [30,31,32,33,74,75].
2.17. Sd/let-7g
The let-7g miRNA belongs to the let-7 family of miRNAs, all of which have been well studied in the context of human cancer. Reviews published in 2010, 2012, and 2017 detail the let-7 family as well as let-7g specifically [86,87,88]. One example from this includes an early publication from 2008 by Kumar et al. where let-7g was found to suppress the NSCLC phenotype both in vitro and in vivo through the RAS pathway [34]. Many publications implicating sd/let-7g as a tumor-suppressing RNA in cancer have emerged since the aforementioned review, including a 2019 publication by Chang et al. in colorectal cancer and a 2019 paper by Biamonte et al. in ovarian cancer [35,36].
2.18. Sd/miR-28
Sd/miR-28 has been identified as a tumor-suppressing RNA in several cancers, including a 2014 publication by Schneider et al. focused on B-cell lymphoma (BCL) [37]. The sdRNA was downregulated in patient BCL samples (n = 25) compared to normal B cells (n = 4), and sd/miR-28 overexpression in BCL cell lines suppressed proliferation and colony formation while enhancing apoptosis. Target engagement was confirmed for sd/miR-28 with the 3′-UTRs of MAD2L1, BAG1, RAP1B, and RAB23 mRNA. Taken together, sd/miR-28 suppresses the BCL phenotype by regulating the expression of four oncogenes involved in cell cycle progression and apoptosis.
2.19. pi-sno74, pi-sno75, pi-sno44, pi-sno78, and pi-sno81
Small RNA-seq (smRNAseq) of patient breast cancer tissue identified five differentially expressed sno-derived piRNAs (pi-snos) within the long noncoding RNA (lncRNA) GAS5 locus [76]. These pi-sno’s included pi-sno74, pi-sno75, pi-sno44, pi-sno78, and pi-sno81. Each of these were found to be downregulated in breast cancer compared to adjacent normal tissue. Microarray and PCR revealed that pi-sno75 upregulates expression of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), a proapoptotic protein. In silico target prediction identified a locus of 169 basepairs (bp) upstream of the TRAIL transcription start site that was highly complementary to pi-sno75 with predicted thermodynamically stable binding. To further investigate this mechanism of action, knockout (KO) and Co-IPs were performed to identify PIWIL1/4 binding partners during pi-sno75 overexpression. This revealed that pi-sno75/PIWIL can specifically interact with the methyltransferase complex protein WDR5, thereby increasing TRAIL expression. In MCF7 cells, pi-sno75 overexpression greatly increased TRAIL expression and, in combination with doxorubicin treatment, enhanced apoptosis. Treatment with pi-sno75 alone resulted in marked reductions of tumor volume in MCF7 and MDA-MB-231 mouse xenografts. In summary, the sno-derived piRNA pi-sno75 recruits epigenetic machinery to specifically upregulate TRAIL, and thus functions as a tumor-suppressive sdRNA.
2.20. pi-sno78 (Sd78-3′)
In 2011, a study by Martens-Uzunova et al. that focused on identifying a miRNA expression signature of prostate cancer also identified significantly differentially expressed sdRNAs [89]. While the majority of reads aligned to defined microRNAs, sdRNAs were found to be predominantly upregulated in the metastatic LN-PCa compared to local prostate cancer. The authors then focused on canonical miRNAs, leaving these enticing putative sdRNA drivers of metastatic LN-PCa uncharacterized until their follow-up publication in 2015. In their 2015 publication, Martens-Uzunova et al. specifically focused on investigating the role of sdRNAs in prostate cancer progression [77]. Consistent with their 2011 study, they identified an sdRNA from the 3′ end of snoRNA78 “sd78-3′” that was upregulated in the more aggressive LN-PCa patient samples. By examining expression at multiple stages of PCa, the authors concluded that globally overexpressed sdRNAs, including sd78-3′, are already present at early stages of cancer but exhibit striking overexpression concordantly with malignant transformation. While the authors did not investigate the mechanism of action of sd78-3′, this sdRNA is actually the pi-sno78 characterized in the above study by He et al. [76].
2.21. piR-017061
Revisiting the 2015 publication by Muller et al. which queried small RNA expression in PDAC patient samples and normal pancreas controls, a total of 123 ncRNAs were found to be significantly differentially expressed [70]. This approach considered 45 noncoding RNAs identified as significantly down-regulated in PDAC, of which there were fourteen sdRNAs and a single sno-derived piRNA, piR-017061.
This entry is adapted from the peer-reviewed paper 10.3390/biomedicines10081819
This entry is offline, you can click here to edit this entry!
