Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Maize is one of the most important crops for human and animal consumption and contains a chemical arsenal essential for survival: flavonoids. Moreover, flavonoids are well known for their beneficial effects on human health. A total of twenty-one genes for the flavonoid pathway of maize was described. The first three genes participate in the general phenylpropanoid pathway. Four genes are common biosynthetic early genes for flavonoids, and fourteen are specific genes for the flavonoid subgroups, the anthocyanins, and flavone C-glycosides.
- Zea mays L.
- anthocyanins
- biosynthesis
- flavonoids
1. Introduction
The comprehension of the maize flavonoid pathways is necessary for plant breeders who want to develop new pigmented maize varieties with better nutraceutical properties and for any health and food scientists working with phenolic compounds. The diversity in the palette of color in maize seeds correlates with differences in the pigment, including carotenoids and flavonoids. Researchers will deepen into these aspects to explain the impressive correlation between plant color, plant survival, and human health.
In maize (Zea mays L.), flavonoids act as deterrents against herbivores, regulate pollen development, and have defensive roles against UV-B radiation [1][2][3]. Flavonoids are a large family of phenolic compounds that share a biosynthetic pathway and, therefore, a common chemical arrangement. The basic structure consists of a C15 skeleton arranged in a C6-C3-C6 where one of the C6 corresponds to a phenyl that is bound to a benzopyran (C6-C3) denominated chromene according to the IUPAC nomenclature (Figure 1). Flavonoids originate from the mevalonate and phenylpropanoid pathways converging in C6-C3-C6 compounds. Some flavonoid molecules differentiate themselves by the chemical changes in the pyran ring, also known as the flavonoid’s C ring [4][5]. For example, anthocyanidins have a modified benzopyran structure with a double bond between the oxygen atom and C2, forming the flavylium cation. Meanwhile, flavones have a double bond between carbons 2 and 3 and a carbonyl group at the C4 position.
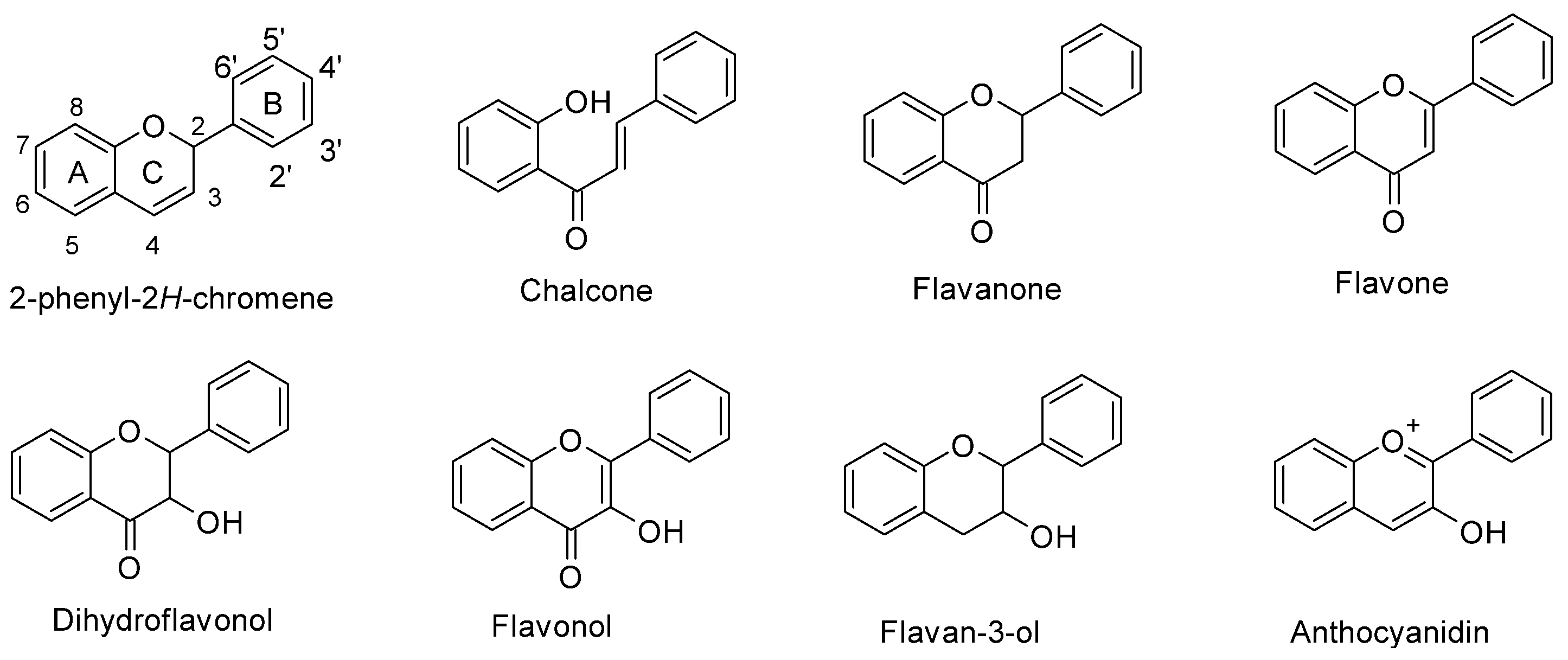 Figure 1. Chemical structure of flavonoid subgroups and the basic C6-C3-C6 skeleton (2-phenyl-2H-chromene). A, B, and C refer to a specific ring of the flavonoid skeleton.
Figure 1. Chemical structure of flavonoid subgroups and the basic C6-C3-C6 skeleton (2-phenyl-2H-chromene). A, B, and C refer to a specific ring of the flavonoid skeleton.2. Structural Protein Genes of the Maize Flavonoid Pathway
2.1. Phenylpropanoid Pathway
The first enzymatic steps in the flavonoid pathway are from three genes of the phenylpropanoid pathway (Table 1). These three enzymes direct the transformation of phenylalanine to coumaroyl-CoA. Those genes are ZmPAL (phenylalanine ammonium lyase, multiples genes, EC 4.3.1.24) [6], ZmC4H (cinnamic acid 4-hydroxylase, Zm00001d009858, EC 1.14.14.91) [6][7], and Zm4CL (4-coumarate CoA ligase, bm5, EC 6.2.1.12) [8]. The three genes share a similar expression profile of downstream genes in the flavonoid pathway in anthocyanin-pigmented tissues [9][10]. Recent analyses have demonstrated multiple gene families in flavonoid biosynthesis with a tissue-specific expression. In addition, some genes such as Zm4CL codify various isoforms, each of which has specific functions [11]. The research on these genes focuses on their roles in lignin biosynthesis [8]. For example, under sugarcane mosaic virus (SCMV) infection, ZmPAL and ZmC4H genes are upregulated, generating the substrate for lignin production [6]. Meanwhile, studies on a brown midrib5 maize line demonstrated that a Zm4CL mutant was responsible for defective lignin biosynthesis [12]. There is a correlation between anthocyanins and lignin where the fungi Ustilago maydis activates the anthocyanin but reduces the lignin biosynthesis, thus facilitating its invasion into the maize seed [13].
Table 1. Summary of genes involved in the early steps of the maize flavonoid pathway.
| Gene Name | Locus | Enzyme/Protein Name | EC | Reference |
|---|---|---|---|---|
| (ZmPAL) | m* | Phenylalanine ammonium lyase | 4.3.1.24 | [6] |
| (ZmC4H) | 8L | Cinnamic acid 4-hydroxylase | 1.14.14.91 | [6][7] |
| bm5 (Zm4CL) | 5 | 4-Coumarate CoA ligase | 6.2.1.12 | [12] |
| c2 (ZmCHS) | 4L | Chalcone synthase | 2.3.1.74 | [14] |
| whp1 (ZmCHS) | 2L | Chalcone synthase | 2.3.1.74 | [15] |
| chi1 (ZmCHI) | 1L | Chalcone isomerase | 5.5.1.6 | [16] |
| fht1 (ZmF3H) | 2S | Flavonoid 3-dioxygenase | 1.14.11.9 | [17] |
| pr1 (ZmF3′H) | 5L | Flavonoid 3′-monooxygenase | 1.14.14.82 | [18] |
2.2. Early Biosynthetic Genes of Flavonoids
2.2.1. Chalcone Synthase (ZmCHS, c2, EC 2.3.1.74)
The first crucial step in flavonoid biosynthesis (Figure 2) is the production of the naringenin chalcone (C6-C3-C6) from the condensation of three molecules of malonyl-CoA (3 × C2) using a 4-coumaroyl-CoA (C6-C3) as substrate [21]. This gene is also known as polyketide synthase (PKS) type III. The chalcone synthase (CHS) works similarly to other PKS enzymes from the mevalonate/acetate pathway [4]. The reaction extends the aliphatic chain from the coumaroyl-CoA three times using two carbon units from a malonyl-CoA. Then, an intramolecular Claisen condensation occurs to form the second aromatic ring.
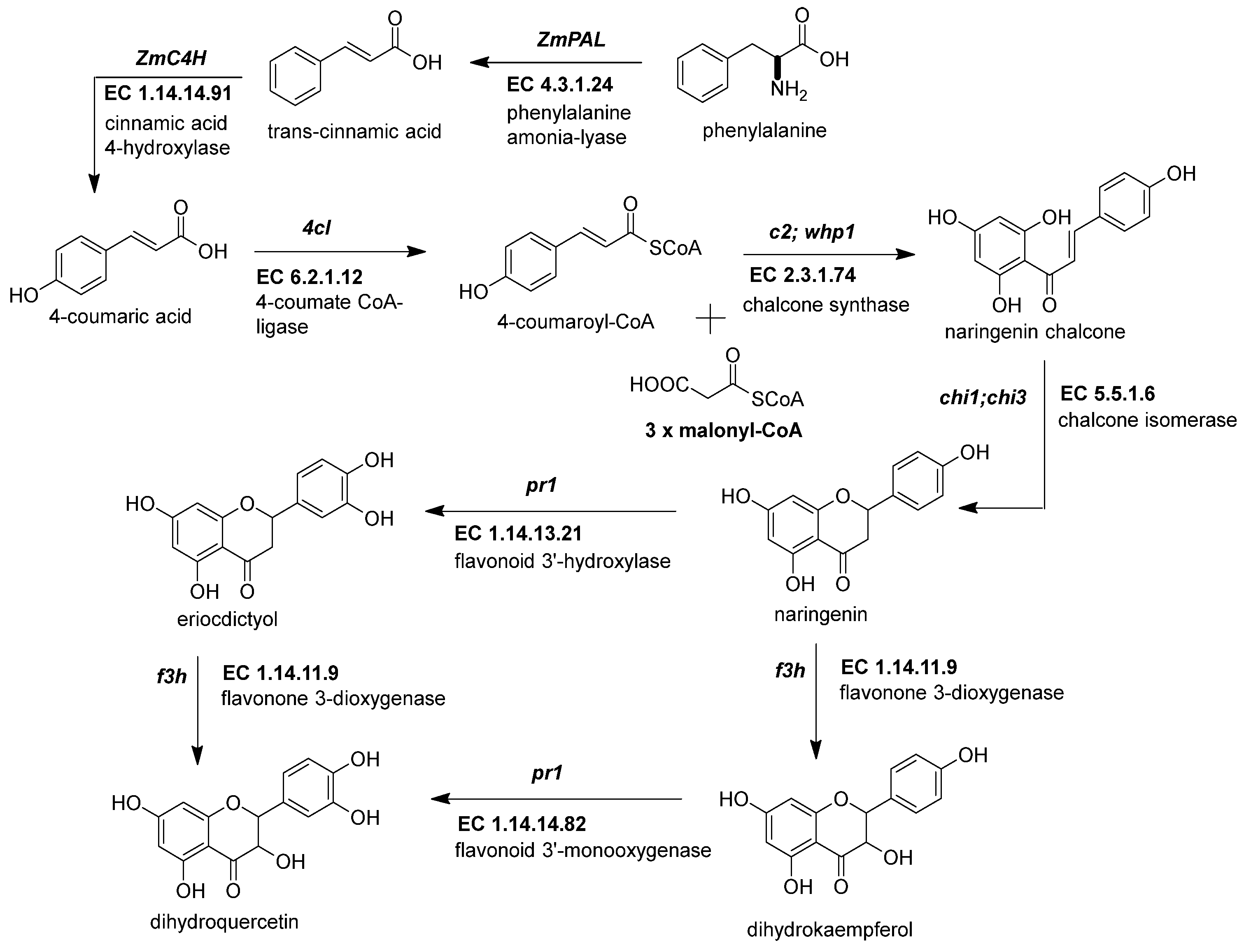 Figure 2. Early genes in the flavonoid pathway. The flavonoid pathway begins with the transformation of phenylalanine to coumaroyl-CoA. The last steps end with the intravacuolar accumulation of acylated anthocyanins. The genes responsible for supplying the coumaroyl-CoA into the flavonoid pathway are phenylalanine ammonium lyase (ZmPAL, EC 4.3.1.24), cinnamic acid 4-hydroxylase (ZmC4H, EC 1.14.14.91), and 4-coumarate CoA ligase (Zm4CL, bm5, EC 6.2.1.12). The flavonoid genes are divided into early biosynthetic genes (EBGs) and late biosynthetic genes (LBGs). EBGs comprise four genes: chalcone synthase (ZmCHS, c2, EC 2.3.1.74), chalcone isomerase (ZmCHI, chi1, EC 5.5.1.6), flavonoid 3-dioxygenase (ZmF3H, fht1, EC 1.14.11.9), and flavonoid 3′-monooxygenase (ZmF3′H, pr1, EC 1.14.14.82). References: [20][22].
Figure 2. Early genes in the flavonoid pathway. The flavonoid pathway begins with the transformation of phenylalanine to coumaroyl-CoA. The last steps end with the intravacuolar accumulation of acylated anthocyanins. The genes responsible for supplying the coumaroyl-CoA into the flavonoid pathway are phenylalanine ammonium lyase (ZmPAL, EC 4.3.1.24), cinnamic acid 4-hydroxylase (ZmC4H, EC 1.14.14.91), and 4-coumarate CoA ligase (Zm4CL, bm5, EC 6.2.1.12). The flavonoid genes are divided into early biosynthetic genes (EBGs) and late biosynthetic genes (LBGs). EBGs comprise four genes: chalcone synthase (ZmCHS, c2, EC 2.3.1.74), chalcone isomerase (ZmCHI, chi1, EC 5.5.1.6), flavonoid 3-dioxygenase (ZmF3H, fht1, EC 1.14.11.9), and flavonoid 3′-monooxygenase (ZmF3′H, pr1, EC 1.14.14.82). References: [20][22].Genome-wide analysis revealed up to 15 ZmCHS genes in the maize genome (Han et al., 2016). However, the members more consistently studied are the duplicated c2 (ZmCHS01) and whip1 (ZmCHS02) genes (Table 1) [14]. Multiple tissues, including tassels, ear husks, and the aleurone layer of endosperm at different developmental stages, express the genes c2 and whip1 [23][24]. Indeed, functional alleles for genes c2 and whip1 are vital to increasing the biosynthesis of any flavonoids downstream, such as apigenin and tricin, essential for lignin formation, and C-glycosyl flavones [15][25]. Meanwhile, members of the chalcone synthase family, such as ZmCHS013 and ZmCHS014, compared to ZmCHS01, had a lower expression in most tissues and different responses under the stimuli of salicylic acid [26].
2.2.2. Chalcone Isomerase (ZmCHI, chi1, EC 5.5.1.6)
This enzyme catalyzes an intramolecular Michael-type addition from the chalcone 2-O to its α,β-unsaturated carbonyl (Figure 2). The final product is the typical phenyl-chromanone or flavanone structure [4]. The first gene sequenced from this family in maize was ZmCHI (Table 1) [16]. Interestingly, mutants have not been reported in maize for this gene, due to the multiple homologous sequences found for ZmCHI in the maize genome. An experiment designed to find QTLs for resistance to Fusarium corn fungi detected ZmCHI3 as a second member of the family [27]. Indeed, a transformed maize callus with a copy of ZmCHI3 from a resistant inbred was less susceptible to maize plagues.
2.2.3. Flavonoid 3-Dioxygenase (ZmF3H, fht1, EC 1.14.11.9)
ZmF3H is a Fe2+ and 2-oxoglutarate-dependent dioxygenase that introduces a hydroxyl group in position 3 of the chalcone structure, generating a dihydroflavonol [17]. There is just one gene copy known in the maize genome. In a previous report, ZmF3H was found to be the only gene in the flavonoid pathway in which mRNA expression levels correlate with the synthesis of flavonols in anthers [28]. Moreover, its expression increases in pigmented kernels compared to white seeds [9].
2.2.4. Flavonoid 3′-Monooxygenase (ZmF3′H, pr1, EC 1.14.14.82)
This pr1 or purple aleurone1 gene has been studied in maize because its alleles are responsible for changes in color pigmentation caused by a difference in anthocyanin profile [29][30]. ZmF3′H is monooxygenase hydroxylate in the 3′ position from the phenyl ring B (Table 1). When the gene is functional, its enzyme can produce blue/violet-colored anthocyanidins (cyanidin and peonidin). If not, it generates a red/orange mono-hydroxylated pelargonidin [18]. Red kernels are homozygous for the recessive alleles pr1 that do not produce functional enzymes, resulting in the pelargonidin-base anthocyanins predominating over the anthocyanin profile. The dominant Pr1 alleles have a gene dose effect in the purple kernel pigmentation, which means that each Pr1 allele in diploid (vegetative) or triploid (endosperm) tissues increase the cyanidin-base anthocyanins (Figure 2) in the pigmented tissue [31].
Moreover, ZmF3′H has a role in the biosynthesis of 3-deoxyflavonoids and phlobaphene; as occurs with the anthocyanins, the precursor transforms into a di-hydroxylated phenyl ring B compound [32]. A Pr1 allele is essential for the resistance against biotic stress depending on C-glucosyl flavone (maysin) accumulation in salmon-colored silks [33].
2.3. Late Biosynthetic Genes of Maize Anthocyanins
2.3.1. Dihydroflavonol 4-Reductase (ZmDFR, a1, EC 1.1.1.219)
This enzyme converts the dihydroflavonol (or flavanonol) to a flavan-3,4 diol by reducing the 4-carbonyl (Figure 3 and Table 2) [34]. There is a hypothesis that this enzyme has a role in phlobaphene biosynthesis by transforming the 4-carbonyl into flavanones to produce 4-flavan-4-ol [35]. The gene locus of ZmDFR, a1, has been deeply studied for two reasons. The first is its linkage to the sh2 gene, responsible for the shrunken seed phenotype, that made possible the studies on transposable elements and meiotic recombination hotspots in the a1-sh2 interval [36][37]. The second reason is that the gene product is a vital enzyme in the flavonoid pathway, favoring which flavonoid subgroup could be biosynthesized [38]. If there is a functional allele, it can produce anthocyanidins (Figure 3) and phlobaphenes (see Section 2.4.1). However, two copies of a non-functional allele would redirect it to flavanol and flavone biosynthesis [39].
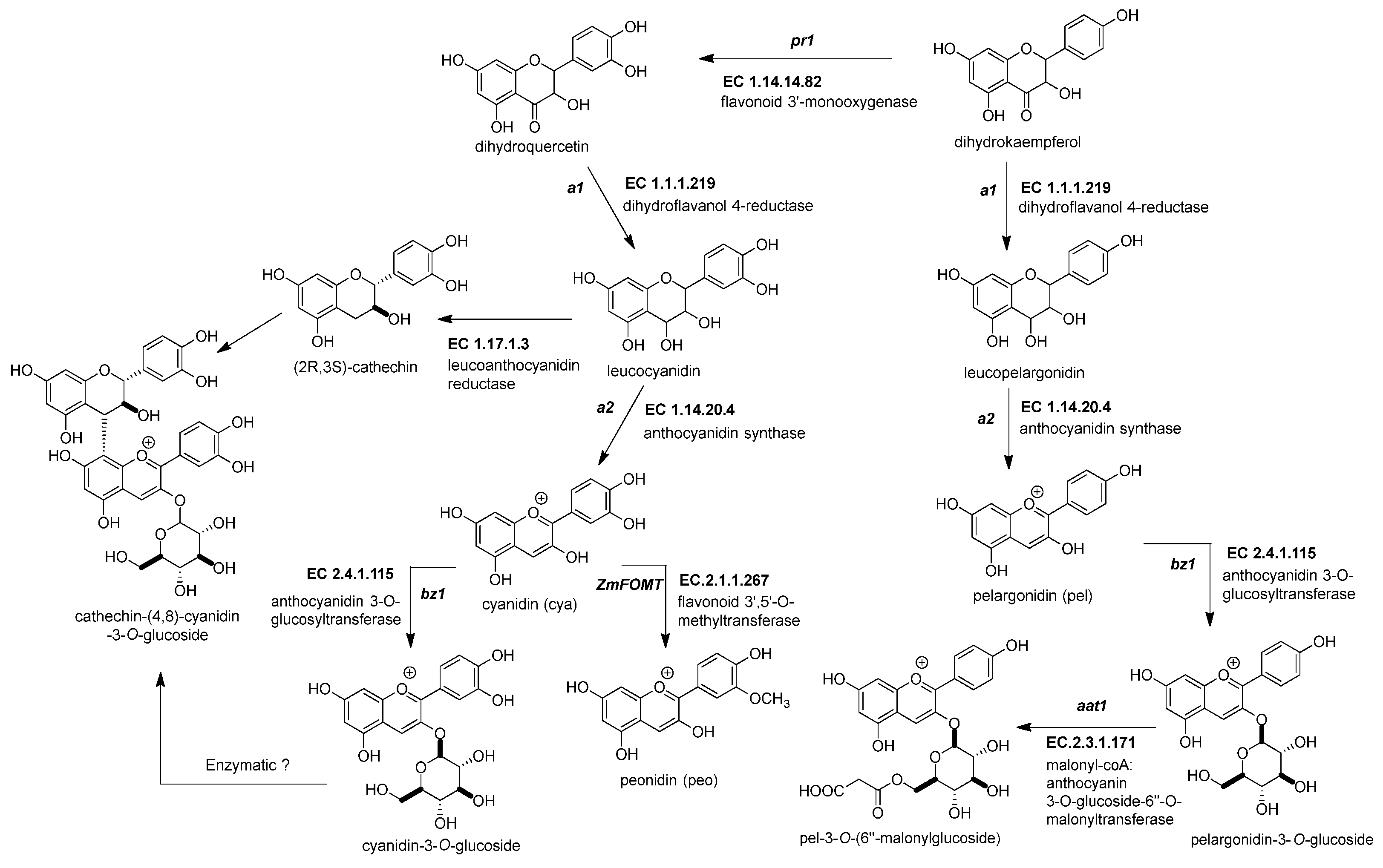 Figure 3. Biosynthetic genes for maize anthocyanin pathway. After the formation of the dihydroflavonol, five enzymatic steps catalyze its biotransformation into acylated maize anthocyanins. Those genes are the following: dihydroflavonol 4-reductase (ZmDFR, a1, EC 1.1.1.219), anthocyanidin synthase (ZmANS, a2, EC 1.14.20.4), anthocyanidin 3-O-glucosyltransferase (ZmAGT, bz1, EC 2.4.1.115), malonyl-CoA: anthocyanin 3-O-glucoside-6′′-O-malonyltransferase (Zm3MAT, aat1, EC 2.3.1.171), and flavonoid 3′,5′-O-methyltransferase (ZmAOMT, EC 2.1.1.267). The glutathione S-transferase (ZmGST, bz2, EC 2.5.1.18) and multidrug resistance protein (ZmABCC3 and ZmABCC4, MRP3 and MRP 4, EC 7.6.2.2) are required to deliver them inside the vacuole. References: [20][22].
Figure 3. Biosynthetic genes for maize anthocyanin pathway. After the formation of the dihydroflavonol, five enzymatic steps catalyze its biotransformation into acylated maize anthocyanins. Those genes are the following: dihydroflavonol 4-reductase (ZmDFR, a1, EC 1.1.1.219), anthocyanidin synthase (ZmANS, a2, EC 1.14.20.4), anthocyanidin 3-O-glucosyltransferase (ZmAGT, bz1, EC 2.4.1.115), malonyl-CoA: anthocyanin 3-O-glucoside-6′′-O-malonyltransferase (Zm3MAT, aat1, EC 2.3.1.171), and flavonoid 3′,5′-O-methyltransferase (ZmAOMT, EC 2.1.1.267). The glutathione S-transferase (ZmGST, bz2, EC 2.5.1.18) and multidrug resistance protein (ZmABCC3 and ZmABCC4, MRP3 and MRP 4, EC 7.6.2.2) are required to deliver them inside the vacuole. References: [20][22].Table 2. Summary of anthocyanin genes in the maize flavonoid pathway.
| Gene Name | Locus | Enzyme/Protein Name | EC | Reference |
|---|---|---|---|---|
| pr1 (ZmF3′H) | 5L | Flavonoid 3′-monooxygenase | 1.14.14.82 | [18] |
| a1 (ZmDFR) | 3L | Dihydroflavonol 4-reductase | 1.1.1.219 | [34] |
| -(ZmLAR) | - | Leucoanthocyanidin reductase | 1.17.1.3 | - * |
| a2 (ZmANS) | 5S | Anthocyanidin synthase | 1.14.20.4 | [40] |
| bz1 (ZmAGT) | 9S | Anthocyanidin 3-O-glucosyltransferase | 2.4.1.115 | [41] |
| aat1 (Zm3MAT) | 1L | Malonyl-CoA: anthocyanin 3-O-glucoside-6′′-O-malonyltransferase | 2.3.1.171 | [42] |
| omt1 and omt4- (ZmAOMT) | 4L | Anthocyanin S-adenosyl-l-methionine-dependent O- methyltransferase | 2.1.1.267 | [43][44] |
| bz2 (ZmGST) | 4L | Glutathione-S-transferase | 2.5.1.18 | [45] |
| mrpa3 (ZmABC3) mrpa4 (ZmABC4) | 9S 1S |
Multidrug resistance-associated protein or ATP-binding cassette transporter | 7.6.2.2 | [46] |
The role of ZmDFR in the diversification of flavonoids is further exemplified by its interaction with multiple transcription factors [9][10]. ZmDFR1 has a gene duplication in the maize genome, known as a4. Nevertheless, it is not clear if there is an active protein in the tissue from the genomic sequences alone [34]. Both genes have a higher expression in pigmented kernels than in anthocyanin-less seeds [9].
2.3.2. Anthocyanidin Synthase (ZmANS, a2, EC 1.14.20.4)
The dioxygenase ZmANS oxidizes at the C-3 position of a flavan-3,4 diol, generating a flavan-3,3,4 triol (Figure 3) [17]. After oxidation, two water molecules are removed, producing an anthocyanidin molecule [47]. Moreover, the ZmANS gene expression is upregulated in pigmented kernels compared to white seeds through elements that conserve the promoter region for the MBW complex [9][40]. The a2 is the unique copy known in the maize genome.
2.3.3. Anthocyanidin 3-O-Glucosyltransferase (ZmAGT, bz1, EC 2.4.1.115)
This enzyme is also known as UDP-flavonoid glucosyltransferase (ZmUFGT). It catalyzes the transference of glucose to the C-3 position of anthocyanidins (Figure 3) [48][49]. This locus is named bronze1 since bz1 alleles cannot produce a functional gene product and are responsible for the bronze-colored aleurone [41][50]. Glycosylated anthocyanidins (anthocyanins) accumulate in a vacuole only when the ZmAGT is functional. If not, the anthocyanidins are prone to oxidation, turning into brown pigments in the cell wall [51]. The expression occurs in all anthocyanin pigmented tissue because it contains conserved elements in its promoter, as other genes are upregulated simultaneously by the MYB-bHLH-WD40 (MBW) complex [50][52].
The locus bz1 is located in the intergenic region bz1-stc1, known for the varying copies of transposable elements [53][54]. A relevant study included the first discovery of the first DNA transposable element, the Ac/Ds transposon, that resulted in a Nobel Prize being awarded to Dr. McClintock [55]. The Ds activation by marker Ac produces a chromosome rupture of chromosome 9 short arm region, which was recognized phenotypically by the apparition of bronze-colored spots in the kernel [56].
2.3.4. Malonyl-CoA: Anthocyanin 3-O-Glucoside-6′′-O-Malonyltransferase (Zm3MAT, aat1, EC 2.3.1.171)
Two types of acyl moieties can modify the glycosidic part of the anthocyanins in the Plantae kingdom, aromatic and aliphatic dicarboxylic acids. Zm3MAT (Figure 3) was the first anthocyanin acyltransferase (AAT) discovered not only in maize but also in monocots [42][57]. Zm3MAT is necessary to produce mono-malonylated anthocyanins, the most common type of anthocyanins in the aleurone layer [58]. Zm3MAT was selected as a QTL for the reduced acylation phenotype and then corroborated through a knockout by Mu transposon insertion [42]. Further research showed that Zm3MAT exerts a dimalonyl transferase activity and can utilize both acyl moieties malonyl-CoA and succinyl-CoA, but it is more specific for malonyl-CoA [57]. The spectrum of anthocyanin selectivity ranges from the most preferable to the least preferable as follows: cyanidin-3-O-glucoside, pelargonidin-3-O-glucoside, peonidin-3-O-glucoside, and delphinidin-3-O-glucoside.
2.3.5. Flavonoid 3′,5′-O-Methyltransferase, or Anthocyanin S-Adenosyl-l-Methionine-Dependent O-Methyltransferase (ZmFOMT or ZmAOMT, EC 2.1.1.267)
This enzyme catalyzes the methylation of a hydroxyl group in the -3′ or -5′ position of the 3-hydroxyflavonoid’s phenyl B-ring (Figure 3) [43]. The enzyme uses several flavonoids as substrates, not just anthocyanins. These include aglycone and glycosylated forms of flavonols or anthocyanidins. However, every member has a specific affinity that favors some substrate above others [58]. Unfortunately, in maize, this enzyme has not been characterized yet. However, Chapman and collaborators mentioned two candidate genes, namely omt1 (Zm00001d052841) and omt4 (Zm00001d05284), for anthocyanin O-methyltransferases related to QTLs for peonidin-base anthocyanins [44].
2.3.6. Glutathione-S-Transferase (ZmGST, bz2, EC 2.5.1.18)
The glutathione S-transferase (GST) family in maize includes more than 40 GST gene sequences [59]. This family of enzymes detoxifies cells affected by xenobiotics, such as herbicides, by conjugating a glutathione (GSH) molecule [60][61][62]. After being labeled with glutathione, these molecules are sent out of the cell by an ATP-dependent glutathione conjugate export pump [63]. However, the bz2 gene, a GST type III, is supposed to label the anthocyanin to be recognized by a vacuolar glutathione pump, and then the labeled anthocyanin is transported into the vacuolar lumen [45][63]. Until now, there is no evidence that shows that anthocyanins are conjugated with GSH. However, the role of bz2 in the accumulation of anthocyanins is accepted. Other researchers suggested that this enzyme may function as a carrier protein for vacuolar anthocyanin sequestration [64].
When ZmGST is not functional, the anthocyanins are not transported to the vacuole interior. Then, the intravacuolar pH and environment contribute to maintaining these molecules without degradation [65]. As described for bz1, a maize plant without functional alleles will develop a bronze-colored kernel [51]. ZmGST is upregulated in pigmented tissue because it shares conserved binding sites in the promoter region for the MBW complex interaction, a characteristic shared with other upstream genes in the flavonoid pathway [9][66].
2.3.7. Multidrug Resistance Protein (ZmABCC3 and -4, mrpa3, EC 7.6.2.2)
ZmABCC3 is part of a broader ATP-binding cassette (ABC) superfamily protein containing up to 130 open reading frames [62]. In maize, this superfamily of transmembrane proteins anchored to the cell membrane is highly specialized in expelling xenobiotics from the intracellular environment [67]. However, ZmABCC3 and ZmABCC4 are present in the tonoplast of vegetative tissues and in the aleurone layer, respectively [46].
This protein follows a similar expression profile to other genes related to anthocyanin biosynthesis [9][68]. Recent research in species such as Vitis vinifera and Arabidopsis thaliana shows that their homologous sequences to ZmABBC3 are GSH/anthocyanin co-transporters [69][70].
2.3.8. Flavanol-Anthocyanin Condensed Forms
The flavanol-anthocyanin condensed forms are compounds found in maize; however, there is still no description of a known enzyme producing them [71]. Their biosynthesis starts with the generation of the flavan-3-ol unit (Figure 3). The leucoanthocyanidin reductase (E.C. 1.17.1.3) participates in a reduction reaction in the C-3 position of the leucoanthocyanidin [44][72]. This enzyme is yet unidentified in maize. Then, a linkage occurs between the anthocyanin and the flavan-3-ol, but there is no recognized enzyme for this process (Figure 3). However, it is known that a QTL for the flavanol-anthocyanin condensed form was mapped near the p1 locus [44].
In wine, the presence of flavanol-anthocyanin condensed forms is related to aging. However, in maize, there is evidence of natural formation [71]. The production of flavanol-anthocyanin condensed forms consumes monomeric anthocyanin, therefore reducing the total concentration [57].
2.4. Biosynthesis of Flavonols, Flavones C-Glycosides, and Phlobaphenes in Maize
2.4.1. Flavonol Synthase (ZmFLS1, fls1, EC 1.14.20.6)
The flavonols are important in maize due to their effects on male fertility and UV-B protection [73]. Flavonol synthesis depends on flavanone 3-dioxygenase and flavonol synthase, a Fe2+/2-oxoglutarate dependent dioxygenase (Figure 4 and Table 3). The transcription factors that regulate the expression of anthocyanins and C-flavone glycosylated biosynthetic genes can also upregulate the expression of ZmFLS1 [1][17]. In the maize genome are two copies (ZmFLS1 and ZmFLS2) in tandem in the long arm of chromosome 5. The expression of both enzymes was augmented under UV-B light and in high-altitude landraces compared to the inbred lines through an increased p1 expression [1][3].
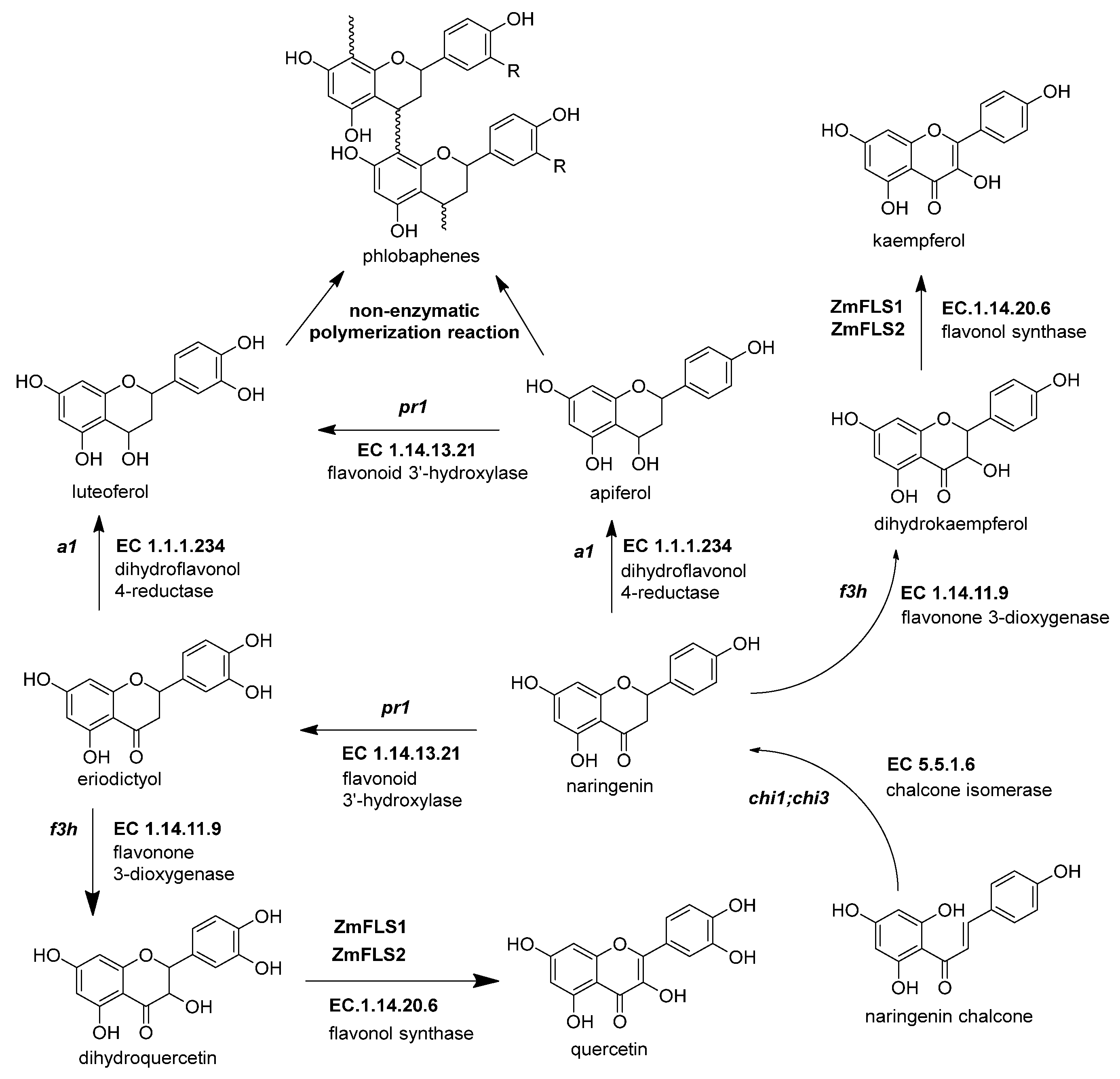 Figure 4. The biosynthetic genes of flavonol and phlobaphenes. The flavanones naringenin and eriodyctiol are the starting substrates for the other flavonoid subgroups. Flavonol synthesis depends on flavanone 3-dioxygenase (ZmF3H, fht1, EC 1.14.11.9) and flavonol synthase (ZmFNS1, fns1, EC 1.14.20.5). Phlobaphene synthesis begins with the action of dihydroflavonol 4-reductase (ZmDFR, a1, EC 1.1.1.219) on flavanones, generating flavan-4-ol molecules that undergo a non-enzymatic polymerization into phlobaphenes. References: [20][22][74].
Figure 4. The biosynthetic genes of flavonol and phlobaphenes. The flavanones naringenin and eriodyctiol are the starting substrates for the other flavonoid subgroups. Flavonol synthesis depends on flavanone 3-dioxygenase (ZmF3H, fht1, EC 1.14.11.9) and flavonol synthase (ZmFNS1, fns1, EC 1.14.20.5). Phlobaphene synthesis begins with the action of dihydroflavonol 4-reductase (ZmDFR, a1, EC 1.1.1.219) on flavanones, generating flavan-4-ol molecules that undergo a non-enzymatic polymerization into phlobaphenes. References: [20][22][74].Table 3. Summary of flavonol and flavone C-glycoside genes in the maize flavonoid pathway.
| Gene Name | Locus | Enzyme/Protein Name | EC | Reference |
|---|---|---|---|---|
| fls1 (ZmFLS1) fls2 (ZmFLS2) |
5L 5L |
Flavonol synthase | 1.14.20.6 | [1] |
| fnsi1 (ZmFNSI1) fnsi2 (ZmFNSI2) |
1S 1S |
Flavone synthase I | 1.14.20.5 | [75] |
| fnsii1 (ZmFNSII1) | 10L | Flavone synthase II | 1.14.19.76 | [2] |
| fns1 (ZmF2H1) | 9L | Flavanone 2-hydroxylase | 1.14.14.162 | [76] |
| cgt1 (ZmCGT) | 6L | UDP-glucose:2-hydroxyflavanone C-glucosyltransferase | 2.4.1.360 | [77] |
| sm2 (UGT91L1) | 2L | flavonol-3-O-glucoside L-rhamnosyltransferase | 2.4.1.159 | [78] |
| sm1 (ZmRHS1) | 6L | Glucose-4,6 dehydratase | 4.2.1.76 | [33] |
2.4.2. Flavone Synthase I (ZmFNSI1-2, fnsi1, EC 1.14.20.5) and Flavone Synthase II (ZmFNSII-1, fnsii1, EC 1.14.19.76)
Maize possesses three enzymes that can synthesize flavones from a flavanone, flavone synthases I and II, and flavone 2-hydroxylase (Figure 5) [2][77]. The flavone synthase produces a desaturation in the C2–C3 bond in the flavanone through an oxidation reaction. The oxidative mechanism in ZmFNSI is a Fe2+/2-oxoglutarate-dependent dioxygenase, like in ZmFLS1, whereas that in ZmFNSII is CYP450 [2]. In addition, ZmFNSI1 is upregulated more in tassels than in silks compared to ZmF2H [79]. The p1 transcription factor regulates the expression of ZmFNSI. Meanwhile, the anthocyanin MBW complex regulates the expression of ZmFNSII. Both types of flavone synthases generate apigenin, which defends the plant against UV-B radiation-induced damage [2].
2.4.3. Flavanone 2-Hydroxylase (ZmF2H1, fns1, EC 1.14.14.162)
In maize, this is the third known enzyme that can produce the flavone backbone of the flavone C-glycosides in the salmon-colored silks [76]. This enzyme is phylogenetically closer to FNS type II, both being CYP proteins [77][80]. Flavanone-2-hydroxylase adds a hydroxyl group into the flavanone C-2, producing the opening of the C-ring and finally generating the 3-oxo-dihydrochalcone (Figure 5). After this opening, it can be glycosylated in either of the two positions of the A-ring, closing the C-ring, eliminating water (spontaneous or not), and then generating in vitro a mixture of C-6 or C-8 glycosylated flavones [77].
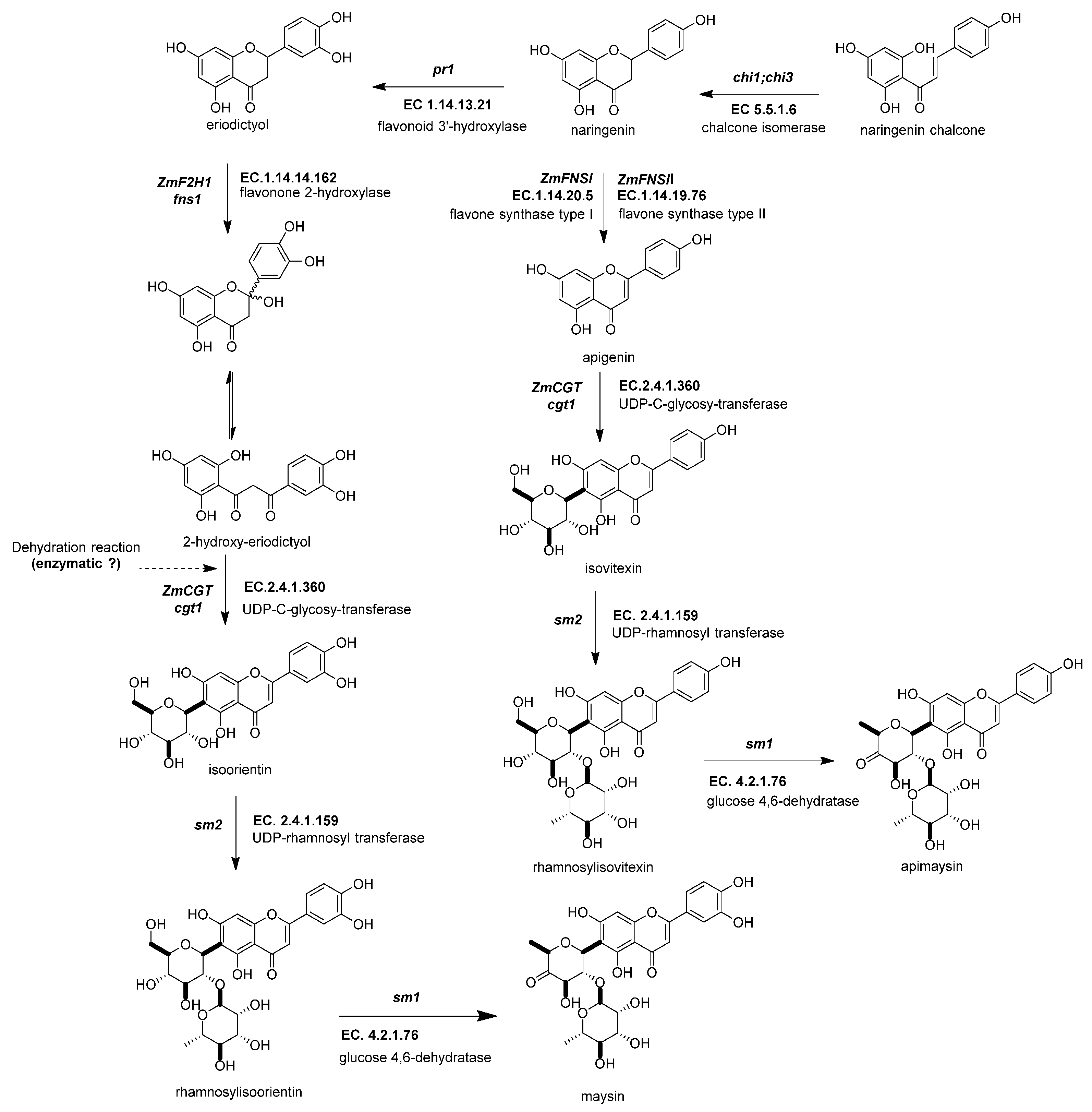 Figure 5. Biosynthetic genes of flavone C-glycosides. The flavanones naringenin and eriodictyol are the initial substrates for the other flavonoid subgroups. There are two possible ways to generate C-glycosyl flavones, indirectly or directly, from any flavanone. The indirect pathway begins through flavanone-2-hydroxylase (ZmF2H, fnsii1, EC 1.14.14.162) opening the C-ring, producing a 3-oxo-dihydrochalcone. Then, UDP C-glycosyl transferase (ZmCGT, cgt1, EC 2.4.1.360) generates a glycosidic bond in the A-ring. Then, there is a dehydration reaction (spontaneous or enzymatic) that produces the C6-flavone glycoside. The direct pathway firstly involves flavone synthase I (ZmFNSII-2, fnsii2, EC 1.14.20.5) and flavone synthase II (ZmFNSII-1, fnsi2, EC 1.14.19.76) producing the same reaction by the addition of a double bond between C2 and C3 in the flavanone. Then, a flavone functions as a substrate for the UDP C-glycosyl transferase (ZmCGT, cgt1, EC 2.4.1.360). The enzymatic action of UDP-rhamnosyl transferase (ZmCGT, sm2, EC 2.4.1.159) and glucose 4,6 dehydratase (sm1, EC 4.2.1.76) produces either apimaysin or maysin. References: [20][33][81].
Figure 5. Biosynthetic genes of flavone C-glycosides. The flavanones naringenin and eriodictyol are the initial substrates for the other flavonoid subgroups. There are two possible ways to generate C-glycosyl flavones, indirectly or directly, from any flavanone. The indirect pathway begins through flavanone-2-hydroxylase (ZmF2H, fnsii1, EC 1.14.14.162) opening the C-ring, producing a 3-oxo-dihydrochalcone. Then, UDP C-glycosyl transferase (ZmCGT, cgt1, EC 2.4.1.360) generates a glycosidic bond in the A-ring. Then, there is a dehydration reaction (spontaneous or enzymatic) that produces the C6-flavone glycoside. The direct pathway firstly involves flavone synthase I (ZmFNSII-2, fnsii2, EC 1.14.20.5) and flavone synthase II (ZmFNSII-1, fnsi2, EC 1.14.19.76) producing the same reaction by the addition of a double bond between C2 and C3 in the flavanone. Then, a flavone functions as a substrate for the UDP C-glycosyl transferase (ZmCGT, cgt1, EC 2.4.1.360). The enzymatic action of UDP-rhamnosyl transferase (ZmCGT, sm2, EC 2.4.1.159) and glucose 4,6 dehydratase (sm1, EC 4.2.1.76) produces either apimaysin or maysin. References: [20][33][81].2.4.4. UDP-Glucose:2-Hydroxyflavanone C-Glucosyltransferase (ZmCGT, cgt1, EC 2.4.1.360)
UDP-glucose:2-hydroxyflavanone C-glucosyltransferase generates a glycosidic bond in the A-ring from the C-1 of the glucose to the C-6 in the C-glycosyl flavones (Figure 5) [33]. In vitro and in vivo experimental evidence has demonstrated that the ZmCGT enzyme has a bifunctional capacity to form glycosidic bonds with C or O atoms. On the contrary, there is only in vitro evidence for C-8 flavone glycosides [76]. The likely reason for that is the possibility of an enzyme that only selects C-6 glycosylated 2-hydroxyflavanone for dehydration into C-6 glycosyl flavones [82].
2.4.5. UDP-Rhamnosyl Transferase (sm2, UGT91L1, EC 2.4.1.159)
The UDP-rhamnosyl transferase enzyme forms the glycosidic bond between the glucose C-2 and the rhamnose C-1 (Figure 5) [78]. Functional alleles confer a characteristic salmon color to the silks due to the accumulation of maysin/apimaysin in the silks. This is due to p1 upregulating sm2 and is expressed principally in silks [33] but also in non-vegetative tissues such as pollen, tassels, and seeds [79].
2.4.6. Glucose-4,6 Dehydratase (ZmRHS1, sm1, EC 4.2.1.76)
The biosynthesis of C-flavones glycosides in maize ends with a modification to the glucose structure of the rhamnosylisoorientin (or rhamnosylisovitexin) to produce maysin/apimaysin (Figure 5) [83]. These metabolites give the ear of maize the ability to deter the herbivore Helicoverpa zea, commonly known as corn earworm [15][33]. This locus was found to be responsible for producing the last step in the maize flavone pathway and found to be a putative UDP-rhamnose synthase (ZmRHS1) [33][78]. The gene has two putative domains; the first domain is a UDP-glucose dehydratase, and the second domain corresponds to UDP glucose 4-keto-6-deoxyglucose epimerase/reductase. The former domain is the exclusive one catalyzing maysin or apimaysin biosynthesis [33]. Its gene expression pattern in the tissues is similar to the sm1 profile [2].
This entry is adapted from the peer-reviewed paper 10.3390/molecules27165166
References
- Falcone Ferreyra, M.L.; Casas, M.I.; Questa, J.I.; Herrera, A.L.; DeBlasio, S.; Wang, J.; Jackson, D.; Grotewold, E.; Casati, P. Evolution and Expression of Tandem Duplicated Maize Flavonol Synthase Genes. Front. Plant Sci. 2012, 3, 101.
- Righini, S.; Rodriguez, E.J.; Berosich, C.; Grotewold, E.; Casati, P.; Falcone Ferreyra, M.L. Apigenin Produced by Maize Flavone Synthase I and II Protects Plants against UV-B-Induced Damage. Plant Cell Environ. 2019, 42, 495–508.
- Rius, S.P.; Emiliani, J.; Casati, P. P1 Epigenetic Regulation in Leaves of High Altitude Maize Landraces: Effect of UV-B Radiation. Front. Plant Sci. 2016, 7, 523.
- Dewick, P.M. Medicinal Natural Products: A Biosynthetic Approach, 3rd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2009; ISBN 9780470742761.
- Rauter, A.P.; Ennis, M.; Hellwich, K.-H.; Herold, B.J.; Horton, D.; Moss, G.P.; Schomburg, I. Nomenclature of Flavonoids (IUPAC Recommendations 2017). Pure Appl. Chem. 2018, 90, 1429–1486.
- Yuan, W.; Jiang, T.; Du, K.; Chen, H.; Cao, Y.; Xie, J.; Li, M.; Carr, J.P.; Wu, B.; Fan, Z.; et al. Maize Phenylalanine Ammonia-Lyases Contribute to Resistance to Sugarcane Mosaic Virus Infection, Most Likely through Positive Regulation of Salicylic Acid Accumulation. Mol. Plant Pathol. 2019, 20, 1365–1378.
- Ôba, K.; Conn, E.E. Induction of Cinnamic Acid 4-Hydroxylase in Developing Maize Seedlings. Phytochemistry 1988, 27, 2447–2450.
- Yun, M.-S.; Chen, W.; Deng, F.; Yogo, Y. Differential Properties of 4-Coumarate: CoA Ligase Related to Growth Suppression by Chalcone in Maize and Rice. Plant Growth Regul. 2005, 46, 169–176.
- Hu, C.; Li, Q.; Shen, X.; Quan, S.; Lin, H.; Duan, L.; Wang, Y.; Luo, Q.; Qu, G.; Han, Q.; et al. Characterization of Factors Underlying the Metabolic Shifts in Developing Kernels of Colored Maize. Sci. Rep. 2016, 6, 35479.
- Li, T.; Zhang, W.; Yang, H.; Dong, Q.; Ren, J.; Fan, H.; Zhang, X.; Zhou, Y. Comparative Transcriptome Analysis Reveals Differentially Expressed Genes Related to the Tissue-Specific Accumulation of Anthocyanins in Pericarp and Aleurone Layer for Maize. Sci. Rep. 2019, 9, 2485.
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four Isoforms of Arabidopsis Thaliana 4-Coumarate: CoA Ligase (4CL) Have Overlapping yet Distinct Roles in Phenylpropanoid Metabolism. Plant Physiol. 2015, 169, 838.
- Xiong, W.; Wu, Z.; Liu, Y.; Li, Y.; Su, K.; Bai, Z.; Guo, S.; Hu, Z.; Zhang, Z.; Bao, Y.; et al. Mutation of 4-Coumarate: Coenzyme A Ligase 1 Gene Affects Lignin Biosynthesis and Increases the Cell Wall Digestibility in Maize Brown Midrib5 Mutants. Biotechnol. Biofuels 2019, 12, 82.
- Tanaka, S.; Brefort, T.; Neidig, N.; Djamei, A.; Kahnt, J.; Vermerris, W.; Koenig, S.; Feussner, K.; Feussner, I.; Kahmann, R. A Secreted Ustilago Maydis Effector Promotes Virulence by Targeting Anthocyanin Biosynthesis in Maize. eLife 2014, 3, e01355.
- Franken, P.; Niesbach-Klösgen, U.; Weydemann, U.; Maréchal-Drouard, L.; Saedler, H.; Wienand, U. The Duplicated Chalcone Synthase Genes C2 and Whp (White Pollen) of Zea Mays Are Independently Regulated; Evidence for Translational Control of Whp Expression by the Anthocyanin Intensifying Gene In. EMBO J. 1991, 10, 2605–2612.
- Szalma, S.J.; Snook, M.E.; Bushman, B.S.; Houchins, K.E.; McMullen, M.D. Duplicate Loci as QTL: The Role of Chalcone Synthase Loci in Flavone and Phenylpropanoid Biosynthesis in Maize. Crop Sci. 2002, 42, 1679–1687.
- Grotewold, E.; Peterson, T. Isolation and Characterization of a Maize Gene Encoding Chalcone Flavonone Isomerase. MGG Mol. Gen. Genet. 1994, 242, 1–8.
- Cheng, A.X.; Han, X.J.; Wu, Y.F.; Lou, H.X. The Function and Catalysis of 2-Oxoglutarate-Dependent Oxygenases Involved in Plant Flavonoid Biosynthesis. Int. J. Mol. Sci. 2014, 15, 1080–1095.
- Sharma, M.; Cortes-Cruz, M.; Ahern, K.R.; McMullen, M.; Brutnell, T.P.; Chopra, S. Identification of the Pr1 Gene Product Completes the Anthocyanin Biosynthesis Pathway of Maize. Genetics 2011, 188, 69–79.
- Jeske, L.; Placzek, S.; Schomburg, I.; Chang, A.; Schomburg, D. BRENDA in 2019: A European ELIXIR Core Data Resource. Nucleic Acids Res. 2019, 47, D542–D549.
- Portwood, J.L.; Woodhouse, M.R.; Cannon, E.K.; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Walsh, J.R.; Sen, T.Z.; Cho, K.T.; Schott, D.A.; et al. Maizegdb 2018: The Maize Multi-Genome Genetics and Genomics Database. Nucleic Acids Res. 2019, 47, D1146–D1154.
- Lim, Y.; Go, M.; Yew, W. Exploiting the Biosynthetic Potential of Type III Polyketide Synthases. Molecules 2016, 21, 806.
- KEGG Flavonoid Biosynthesis. Available online: http://www.genome.jp/kegg-bin/show_pathway?ec00941 (accessed on 2 June 2020).
- Coe, E.H.; McCormick, S.M.; Modena, S.A. White Pollen in Maize. J. Hered. 1981, 72, 318–320.
- Della Vedova, C.B.; Lorbiecke, R.; Kirsch, H.; Schulte, M.B.; Scheets, K.; Borchert, L.M.; Scheffler, B.E.; Wienand, U.; Cone, K.C.; Birchler, J.A. The Dominant Inhibitory Chalcone Synthase Allele C2-Idf (Inhibitor Diffuse) From Zea Mays (L.) Acts via an Endogenous RNA Silencing Mechanism. Genetics 2005, 170, 1989–2002.
- Eloy, N.B.; Voorend, W.; Lan, W.; Saleme, M.d.L.S.; Cesarino, I.; Vanholme, R.; Smith, R.A.; Goeminne, G.; Pallidis, A.; Morreel, K.; et al. Silencing CHALCONE SYNTHASE in Maize Impedes the Incorporation of Tricin into Lignin and Increases Lignin Content. Plant Physiol. 2017, 173, 998–1016.
- Han, Y.; Ding, T.; Su, B.; Jiang, H. Genome-Wide Identification, Characterization and Expression Analysis of the Chalcone Synthase Family in Maize. Int. J. Mol. Sci. 2016, 17, 161.
- Dowd, P.F.; Berhow, M.A.; Johnson, E.T. Enhanced Pest Resistance and Increased Phenolic Production in Maize Callus Transgenically Expressing a Maize Chalcone Isomerase -3 like Gene. Plant Gene 2018, 13, 50–55.
- Deboo, G.B.; Albertsen, M.C.; Taylor, L.P. Flavanone 3-Hydroxylase Transcripts and Flavonol Accumulation Are Temporally Coordinate in Maize Anthers. Plant J. 1995, 7, 703–713.
- Larson, R.L.; Bussard, J.B. Microsomal Flavonoid 3′-Monooxygenase from Maize Seedlings. Plant Physiol. 1986, 80, 483–486.
- Larson, R.; Bussard, J.B.; Coe, E.H. Gene-Dependent Flavonoid 3′-Hydroxylation in Maize. Biochem. Genet. 1986, 24, 615–624.
- Ford, R.H. Inheritance of Kernel Color in Corn: Explanations & Investigations. Source Am. Biol. Teach. Publ. By Natl. Assoc. Biol. Teach. 2000, 62, 181–188.
- Sharma, M.; Chai, C.; Morohashi, K.; Grotewold, E.; Snook, M.E.; Chopra, S. Expression of Flavonoid 3′-Hydroxylase Is Controlled by P1, the Regulator of 3-Deoxyflavonoid Biosynthesis in Maize. BMC Plant Biol. 2012, 12, 1.
- Casas, M.I.; Falcone Ferreyra, M.L.; Jiang, N.; Mejía Guerra, M.K.; Rodríguez, E.; Wilson, T.; Engelmeier, J.; Casati, P.; Grotewold, E. Identification and Characterization of Maize Salmon Silks Genes Involved in Insecticidal Maysin Biosynthesis. Plant Cell 2016, 28, 1297–1309.
- Bernhardt, J.; Stich, K.; Schwarz-Sommer, Z.; Saedler, H.; Wienand, U. Molecular Analysis of a Second Functional A1 Gene (Dihydroflavonol 4-Reductase) in Zea Mays. Plant J. 1998, 14, 483–488.
- Yang, F.; Li, W.; Jiang, N.; Yu, H.; Morohashi, K.; Ouma, W.Z.; Morales-Mantilla, D.E.; Gomez-Cano, F.A.; Mukundi, E.; Prada-Salcedo, L.D.; et al. A Maize Gene Regulatory Network for Phenolic Metabolism. Mol. Plant 2017, 10, 498–515.
- Brown, J.; Sundaresan, V. A Recombination Hotspot in the Maize A1 Intragenic Region. Theor. Appl. Genet. 1991, 81, 185–188.
- Yao, H.; Zhou, Q.; Li, J.; Smith, H.; Yandeau, M.; Nikolau, B.J.; Schnable, P.S. Molecular Characterization of Meiotic Recombination across the 140-Kb Multigenic A1-Sh2 Interval of Maize. Proc. Natl. Acad. Sci. USA 2002, 99, 6157–6162.
- McMullen, M.D.; Snook, M.; Lee, E.A.; Byrne, P.F.; Kross, H.; Musket, T.A.; Houchins, K.; Coe, J. The Biological Basis of Epistasis between Quantitative Trait Loci for Flavone and 3-Deoxyanthocyanin Synthesis in Maize (Zea Mays L.). Genome 2001, 44, 667–676.
- Guo, B.Z.; Zhang, Z.J.; Butrón, A.; Widstrom, N.W.; Snook, M.E.; Lynch, R.E.; Plaisted, D. Lost P1 Allele in Sh2 Sweet Corn: Quantitative Effects of P1 and A1 Genes on Concentrations of Maysin, Apimaysin, Methoxymaysin, and Chlorogenic Acid in Maize Silk. J. Econ. Entomol. 2004, 97, 2117–2126.
- Lesnick, M.L.; Chandler, V.L. Activation of the Maize Anthocyanin Gene A2 Is Mediated by an Element Conserved in Many Anthocyanin Promoters. Plant Physiol. 1998, 117, 437–445.
- Furtek, D.; Schiefelbein, J.W.; Johnston, F.; Nelson, O.E. Sequence Comparisons of Three Wild-Type Bronze-1 Alleles from Zea Mays. Plant Mol. Biol. 1988, 11, 473–481.
- Paulsmeyer, M.N.; Brown, P.J.; Juvik, J.A. Discovery of Anthocyanin Acyltransferase1 (AAT1) in Maize Using Genotyping-by-Sequencing (GBS). G3 Genes Genomes Genet. 2018, 8, 3669–3678.
- Hugueney, P.; Provenzano, S.; Verriès, C.; Ferrandino, A.; Meudec, E.; Batelli, G.; Merdinoglu, D.; Cheynier, V.; Schubert, A.; Ageorges, A. A Novel Cation-Dependent O- Methyltransferase Involved in Anthocyanin Methylation in Grapevine. Plant Physiol. 2009, 150, 2057–2070.
- Chatham, L.A.; Juvik, J.A. Linking Anthocyanin Diversity, Hue, and Genetics in Purple Corn. G3 Genes Genomes Genet. 2021, 11, jkaa062.
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A Glutathione S-Transferase Involved in Vacuolar Transfer Encoded by the Maize Gene Bronze-2. Nature 1995, 375, 397–400.
- Goodman, C.D.; Casati, P.; Walbot, V. A Multidrug Resistance–Associated Protein Involved in Anthocyanin Transport in Zea Mays. Plant Cell 2004, 16, 1812–1826.
- Wilmouth, R.C.; Turnbull, J.J.; Welford, R.W.D.; Clifton, I.J.; Prescott, A.G.; Schofield, C.J. Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana. Structure 2002, 10, 93–103.
- Ford, C.M.; Boss, P.K.; Hæj, P.B. Cloning and Characterization of Vitis Vinifera UDP-Glucose. Flavonoid 3- O-Glucosyltransferase, a Homologue of the Enzyme Encoded by the Maize Bronze-1 Locus That May Primarily Serve to Glucosylate Anthocyanidins in vivo. J. Biol. Chem. 1998, 273, 9224–9233.
- Fukuchi-Mizutani, M.; Okuhara, H.; Fukui, Y.; Nakao, M.; Katsumoto, Y.; Yonekura-Sakakibara, K.; Kusumi, T.; Hase, T.; Tanaka, Y. Biochemical and Molecular Characterization of a Novel UDP-Glucose:Anthocyanin 3′-O-Glucosyltransferase, a Key Enzyme for Blue Anthocyanin Biosynthesis, from Gentian. Plant Physiol. 2003, 132, 1652–1663.
- Roth, B.A.; Goff, S.A.; Klein, T.M.; Fromm, M.E. C1- and R-Dependent Expression of the Maize Bz1 Gene Requires Sequences with Homology to Mammalian Myb and Myc Binding Sites. Plant Cell 1991, 3, 317–325.
- Rhoades, M.M. The Effect of the Bronze Locus on Anthocyanin Formation in Maize. Am. Nat. 1952, 86, 105–108.
- Zhang, J.; Zhang, S.; Li, H.; Du, H.; Huang, H.; Li, Y.; Hu, Y.; Liu, H.; Liu, Y.; Yu, G.; et al. Identification of Transcription Factors ZmMYB111 and ZmMYB148 Involved in Phenylpropanoid Metabolism. Front. Plant Sci. 2016, 7, 148.
- Wang, Q.; Dooner, H.K. Remarkable Variation in Maize Genome Structure Inferred from Haplotype Diversity at the Bz Locus. Proc. Natl. Acad. Sci. USA 2006, 103, 17644–17649.
- Dooner, H.K.; He, L. Maize Genome Structure Variation: Interplay between Retrotransposon Polymorphisms and Genic Recombination. Plant Cell 2008, 20, 249–258.
- Kass, L.B.; Chomet, P. Barbara McClintock. In Handbook of Maize; Springer: New York, NY, USA, 2009; pp. 17–52. ISBN 9780387778631.
- Jones, R.N. McClintock’s Controlling Elements: The Full Story. Cytogenet. Genome Res. 2005, 109, 90–103.
- Paulsmeyer, M.; Juvik, J. Functional Characterization of an Anthocyanin Dimalonyltransferase in Maize. Molecules 2021, 26, 2020.
- Paulsmeyer, M.; Chatham, L.; Becker, T.; West, M.; West, L.; Juvik, J. Survey of Anthocyanin Composition and Concentration in Diverse Maize Germplasms. J. Agric. Food Chem. 2017, 65, 4341–4350.
- McGonigle, B.; Keeler, S.J.; Lau, S.M.C.; Koeppe, M.K.; O’Keefe, D.P. A Genomics Approach to the Comprehensive Analysis of the Glutathione S-Transferase Gene Family in Soybean and Maize. Plant Physiol. 2000, 124, 1105–1120.
- Neuefeind, T.; Huber, R.; Reinemer, P.; Knäblein, J.; Prade, L.; Mann, K.; Bieseler, B. Cloning, Sequencing, Crystallization and X-ray Structure of Glutathione S-Transferase-III from Zea mays Var. Mutin: A Leading Enzyme in Detoxification of Maize Herbicides. J. Mol. Biol. 1997, 274, 577–587.
- Li, D.; Gao, Q.; Xu, L.; Pang, S.; Liu, Z.; Wang, C.; Tan, W. Characterization of Glutathione S-Transferases in the Detoxification of Metolachlor in Two Maize Cultivars of Differing Herbicide Tolerance. Pestic. Biochem. Physiol. 2017, 143, 265–271.
- Pang, K.; Li, Y.; Liu, M.; Meng, Z.; Yu, Y. Inventory and General Analysis of the ATP-Binding Cassette (ABC) Gene Superfamily in Maize (Zea mays L.). Gene 2013, 526, 411–428.
- Alfenito, M.R.; Souer, E.; Goodman, C.D.; Buell, R.; Mol, J.; Koes, R.; Walbot, V. Functional Complementation of Anthocyanin Sequestration in the Vacuole by Widely Divergent Glutathione S-Transferases. Plant Cell 1998, 10, 1135–1149.
- Sun, Y.; Li, H.; Huang, J.R. Arabidopsis TT19 Functions as a Carrier to Transport Anthocyanin from the Cytosol to Tonoplasts. Mol. Plant 2012, 5, 387–400.
- Larsen, E.S.; Alfenito, M.R.; Briggs, W.R.; Walbot, V. A Carnation Anthocyanin Mutant Is Complemented by the Glutathione S-Transferases Encoded by Maize Bz2 and Petunia An9. Plant Cell Rep. 2003, 21, 900–904.
- Schmitz, G.; Theres, K. Structural and Functional Analysis of the Bz2 Locus of Zea mays: Characterization of Overlapping Transcripts. MGG Mol. Gen. Genet. 1992, 233, 269–277.
- Pang, S.; Duan, L.; Liu, Z.; Song, X.; Li, X.; Wang, C. Co-Induction of a Glutathione-S-Transferase, a Glutathione Transporter and an ABC Transporter in Maize by Xenobiotics. PLoS ONE 2012, 7, e40712.
- Bruce, W.; Folkerts, O.; Garnaat, C.; Crasta, O.; Roth, B.; Bowen, B. Expression Profiling of the Maize Flavonoid Pathway Genes Controlled by Estradiol-Inducible Transcription Factors CRC and P. Curr. Opin. Plant Biol. 2000, 3, 173.
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP Binding Cassette Protein from Grape Berry, Transports Anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854.
- Behrens, C.E.; Smith, K.E.; Iancu, C.V.; Choe, J.-y.; Dean, J.V. Transport of Anthocyanins and Other Flavonoids by the Arabidopsis ATP-Binding Cassette Transporter AtABCC2. Sci. Rep. 2019, 9, 437.
- González-Manzano, S.; Pérez-Alonso, J.J.; Salinas-Moreno, Y.; Mateus, N.; Silva, A.M.S.; de Freitas, V.; Santos-Buelga, C. Flavanol-Anthocyanin Pigments in Corn: NMR Characterisation and Presence in Different Purple Corn Varieties. J. Food Compos. Anal. 2008, 21, 521–526.
- Chatham, L.A.; West, L.; Berhow, M.A.; Vermillion, K.E.; Juvik, J.A. Unique Flavanol-Anthocyanin Condensed Forms in Apache Red Purple Corn. J. Agric. Food Chem. 2018, 66, 10844–10854.
- Ferreyra, M.L.F.; Rius, S.; Emiliani, J.; Pourcel, L.; Feller, A.; Morohashi, K.; Casati, P.; Grotewold, E. Cloning and Characterization of a UV-B-Inducible Maize Flavonol Synthase. Plant J. 2010, 62, 77–91.
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, Biological Functions, and Biotechnological Applications. Front. Plant Sci. 2012, 3, 1–15.
- Falcone Ferreyra, M.L.; Emiliani, J.; Rodriguez, E.J.; Campos-Bermudez, V.A.; Grotewold, E.; Casati, P. The Identification of Maize and Arabidopsis Type I FLAVONE SYNTHASEs Links Flavones with Hormones and Biotic Interactions. Plant Physiol. 2015, 169, 1090–1107.
- Falcone Ferreyra, M.L.; Rodriguez, E.; Casas, M.I.; Labadie, G.; Grotewold, E.; Casati, P. Identification of a Bifunctional Maize C-and O-Glucosyltransferase. J. Biol. Chem. 2013, 288, 31678–31688.
- Vanegas, K.G.; Larsen, A.B.; Eichenberger, M.; Fischer, D.; Mortensen, U.H.; Naesby, M. Indirect and Direct Routes to C-Glycosylated Flavones in Saccharomyces Cerevisiae. Microb. Cell Fact. 2018, 17, 1–10.
- McMullen, M.D. Salmon Silk Genes Contribute to the Elucidation of the Flavone Pathway in Maize (Zea mays L.). J. Hered. 2004, 95, 225–233.
- Zhang, Z.; Liang, Z.; Yin, L.; Li, Q.X.; Wu, Z. Distribution of Four Bioactive Flavonoids in Maize Tissues of Five Varieties and Correlation with Expression of the Biosynthetic Genes. J. Agric. Food Chem. 2018, 66, 10431–10437.
- Zhang, T.; Lv, W.; Zhang, H.; Ma, L.; Li, P.; Ge, L.; Li, G. Genome-Wide Analysis of the Basic Helix-Loop-Helix (BHLH) Transcription Factor Family in Maize. BMC Plant Biol. 2018, 18, 235.
- Morohashi, K.; Casas, M.I.; Falcone Ferreyra, M.L.; Mejía Guerra, M.K.; Pourcel, L.; Yilmaz, A.; Feller, A.; Carvalho, B.; Emiliani, J.; Rodriguez, E.; et al. A Genome-Wide Regulatory Framework Identifies Maize Pericarp Color1 Controlled Genes. Plant Cell 2012, 24, 2745–2764.
- Brazier-Hicks, M.; Evans, K.M.; Gershater, M.C.; Puschmann, H.; Steel, P.G.; Edwards, R. The C -Glycosylation of Flavonoids in Cereals. J. Biol. Chem. 2009, 284, 17926–17934.
- McMullen, M.D.; Byrne, P.F.; Snook, M.E.; Wiseman, B.R.; Lee, E.A.; Widstrom, N.W.; Coe, E.H. Quantitative Trait Loci and Metabolic Pathways. Proc. Natl. Acad. Sci. USA 1998, 95, 1996–2000.
This entry is offline, you can click here to edit this entry!
