Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The lack of highly specific and sensitive biomarkers for the early detection of prostate cancer (PCa) is a major barrier to its management. Volatilomics emerged as a non-invasive, simple, inexpensive, and easy-to-use approach for cancer screening, characterization of disease progression, and follow-up of the treatment’s success.
- prostate cancer
- diagnosis
- volatilomics
1. Introduction
Prostate cancer (PCa) is the second most frequent malignant tumor, the fifth leading cause of cancer death among men worldwide (the leading cause of cancer death among men in 46 countries), and the most frequently diagnosed cancer in 105 of 185 of the world countries [1]. In 2020, almost 1.4 million new cases and about 0.4 million deaths were estimated (GLOBOCAN data) [2]. PCa is very heterogeneous in terms of grade and genetics, displaying complex biological, hormonal, and molecular features [2]. This cancer has different phenotypes, ranging from indolent asymptomatic, a non-life-threatening form, to metastatic, very aggressive, rapidly progressive, and lethal forms [3][4]. Unlike diseases such as breast and colon cancer, no major predisposition genes for PCa have been detected. Instead, multiple chromosomal loci of susceptibility genes have been identified, and most of the genomic regions remain poorly studied, which explains this cancer’s heterogeneity [5]. Furthermore, epigenetic factors play an important role in its clinical phenotypes [6].
PCa and subsequent treatments have a high impact on the functional and psychological status of patients, significantly affecting their quality of life [7]. The current diagnostic methods are based on the measurement of prostate-specific antigen (PSA) blood levels, transrectal ultrasound, digital rectal examinations (DRE), and prostate biopsies [8] (Figure 1). However, these methods are invasive, expensive, and unpleasant to patients, with consequent risks of unnecessary complications, and can lead to both false-positive and false-negative results [9]. The PSA test has limited sensitivity (20.5%) [10], accuracy (62–75%) [11], and specificity (51–91%) [12]. Its low selectivity to detect PCa often leads to the overdiagnosis and overtreatment of relatively indolent tumors with low potential for morbidity or death if left untreated [13].
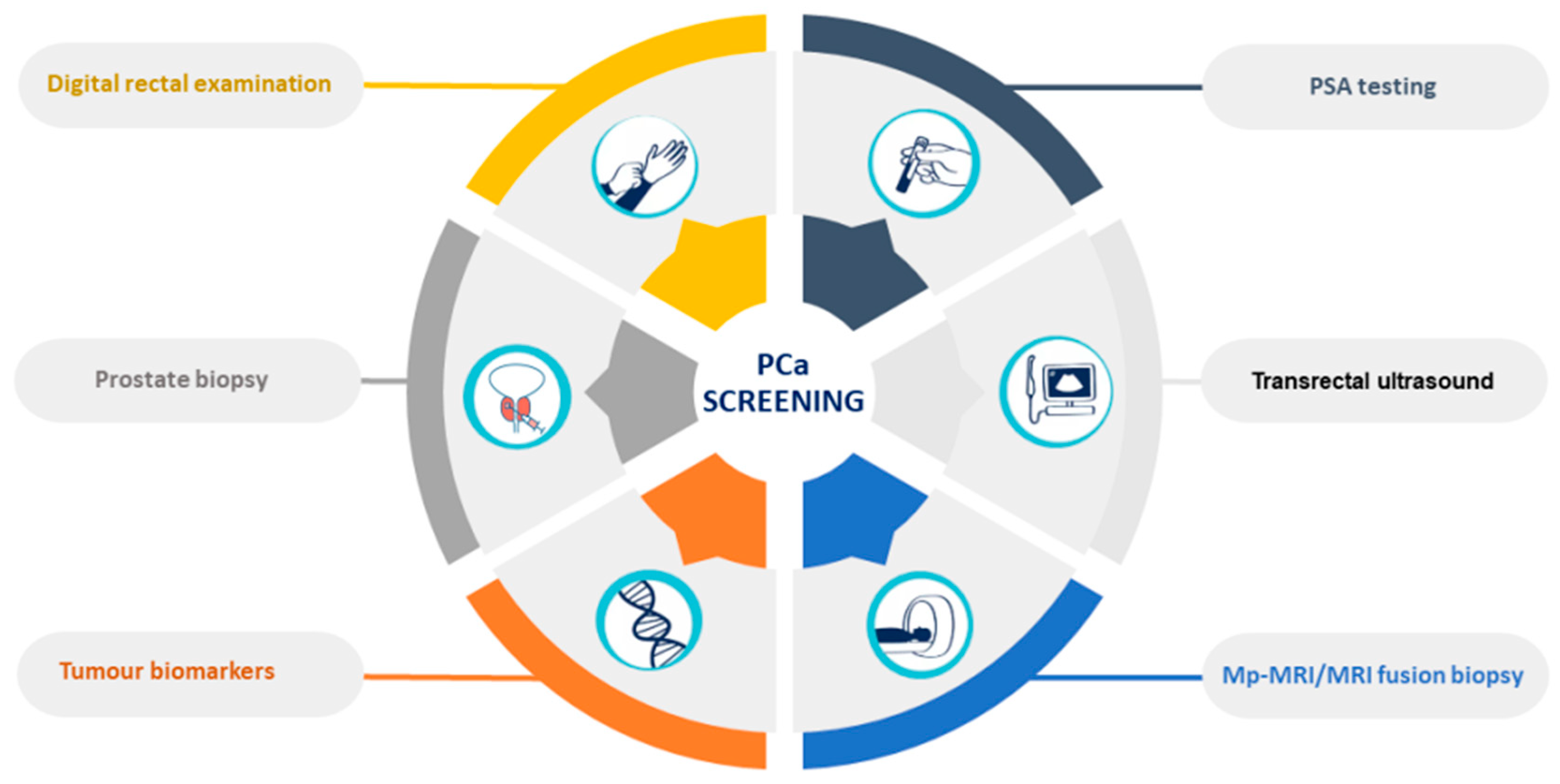
Figure 1. Prostate-cancer screening methods.
The advances in OMICs science have contributed to the discovery of new biomarkers for PCa detection, management, and surveillance. Despite the great efforts and important discoveries, no biomarker has been able to replace PSA in clinical practice for PCa screening [10]. Hence, need is urgent to find highly specific diagnostic tools for non-invasive detection of PCa that are preferentially able to stratify patients by cancer aggressiveness and consequent choice of therapy, which will lead to personalized and targeted therapies. More recently, volatilomics emerged as a promising approach for the definition of cancer biomarkers, based on metabolites biosynthesized by different metabolic pathways, and found in readily accessible biofluids, such as saliva, exhaled breath, and urine.
2. Prostate Cancer Biomarkers
In recent years, advances in molecular medicine have contributed to the discovery of new potential biomarkers to aid in PCa screening and management. Common liquid biopsies biomarkers include extracellular vesicles (EVs), circulating tumor cells (CTCs) and DNA (ctDNA), and cell-free DNA (cfDNA) [14]. However, a few issues prevent the effective use of CTCs and EVs as biomarkers in liquid biopsies for diagnosing PCa, such as the need for specific guidelines for the biomarker’s isolation and detection. Moreover, the microfluidic devices used to develop liquid biopsies have not yet been fully validated and standardized [15]. Long noncoding RNAs (lncRNAs) have also emerged as a promising class of PCa biomarkers. Most lncRNAs associated with PCa are overexpressed in tumor tissues and cancer cells, contributing to tumor proliferation, invasion, and metastasis. In contrast, only a few lncRNAs are downregulated and may act as tumor suppressors, in addition to their potential activity as transcriptional regulators and oncogenes. All of these unique features make lncRNAs promising predictive biomarkers and therapeutic targets for the diagnosis, screening, prognosis, and progression of PCa. Nevertheless, the molecular mechanisms of action of lncRNAs are not very clear yet and it will be important to fully understand and investigate the roles and mechanisms of lncRNAs in prostate carcinogenesis [16]. Other molecular biomarkers for urine, serum, and tissue samples have been developed (Table 1) based on the combination of imaging techniques with other methodologies, such as gene or protein profiling, to enhance cancer detection, pre-biopsy decision, determination of cancer risk, and therapeutic management of PCa [17].
Table 1. Potential clinical utility, characteristics, and availability of prostate cancer biomarkers.
| Biomarker Test | Molecular Markers | Potential Clinical Utility | Characteristics | Availability |
|---|---|---|---|---|
| Serum biomarkers | ||||
| PSA | PSA | Treatment monitoring | Sensitivity: 60% [18] Specificity: 79% [18] AUC: 0.55 [19] |
|
| 4KScore | Total PSA, free PSA, intact PSA, hK2 | Unnecessary biopsy reduction of 43% [20] Risk prediction of PCa metastases Previous negative biopsy |
Sensitivity: 75% [21] Specificity: 63% [21] AUC: 0.71 [22] |
CLIA-certified |
| PHI | Total PSA, free PSA, p2PSA isoform | Unnecessary biopsy reduction of 40% [23] Prediction of high-grade PCa Active supervision monitoring |
Sensitivity: 82% [24] Specificity: 80% [24] AUC: 0.71 [21] |
FDA-approved |
| Urinary biomarkers | ||||
| Progensa (PCA3) | Long non-coding RNAs (ratio of PCA3 mRNA:PSA mRNA) | Unnecessary biopsy reduction of 23–38% [25] PCa detection, staging, and prognosis Previous negative biopsy |
Sensitivity: 69% [26] Specificity: 65% [26] AUC: 0.73 [26] |
FDA-approved |
| SelectMDx | HOXC6 and DLX1 mRNA | Unnecessary biopsy reduction of 53% [27] Prediction of high-grade PCa |
Sensitivity: 91% [27] Specificity: 36% [27] AUC: 0.71–0.81 [27] |
CLIA-certified |
| MPS | PCA3 and TMPRSS2-ERG mRNA | Unnecessary biopsy reduction of 35–47% [28] Predict the risk of PCa and high-grade PCa |
Sensitivity: 93% [29] Specificity: 33% [29] AUC: 0.69 [28] |
CLIA-certified |
| EPI | Exosomal RNA (SPDEF, PCA3, ERG) | Unnecessary biopsy reduction of 27% [30] Improved identification of high-grade PCa |
Sensitivity: 92% [30] Specificity: 34% [30] AUC: 0.70 [30] |
CLIA-certified |
| Tissue biomarkers | ||||
| ConfirmMDx | DNA hypermethylation GSTP1, APC, and RASSF1 | Prediction of true negative prostate biopsies | Sensitivity: 68% [24] Specificity: 64% [24] AUC: 0.74 [24] |
Not FDA-approved yet |
| OncotypeDX | mRNA expression (17 genes) | Monitoring of tumor aggressiveness | AUC: 0.73 [26] | Not FDA-approved yet |
| Prolaris | mRNA expression (31 genes) | Monitoring of tumor aggressiveness | AUC: 0.78 [26] | FDA-approved |
| Decipher | mRNA expression (22 genes) | Treatment monitoring | Sensitivity: 73% [31] Specificity: 74% [31] AUC: 0.79 [31] |
CLIA-certified |
| ProMark | Protein biomarker test (8 proteins) | Monitoring of tumor aggressiveness | Sensitivity: 90% [27] Specificity: 85% [27] AUC: 0.72 [27] |
CLIA-certified |
Abnormalities in these tests indicate the performance of a prostate biopsy. Moreover, risk calculators are combined with these tests to help determine the risk of cancer, thus reducing the number of unnecessary biopsies. The guidelines recommend using these tests in combination with the current PCa screening methods (Figure 2) [10].
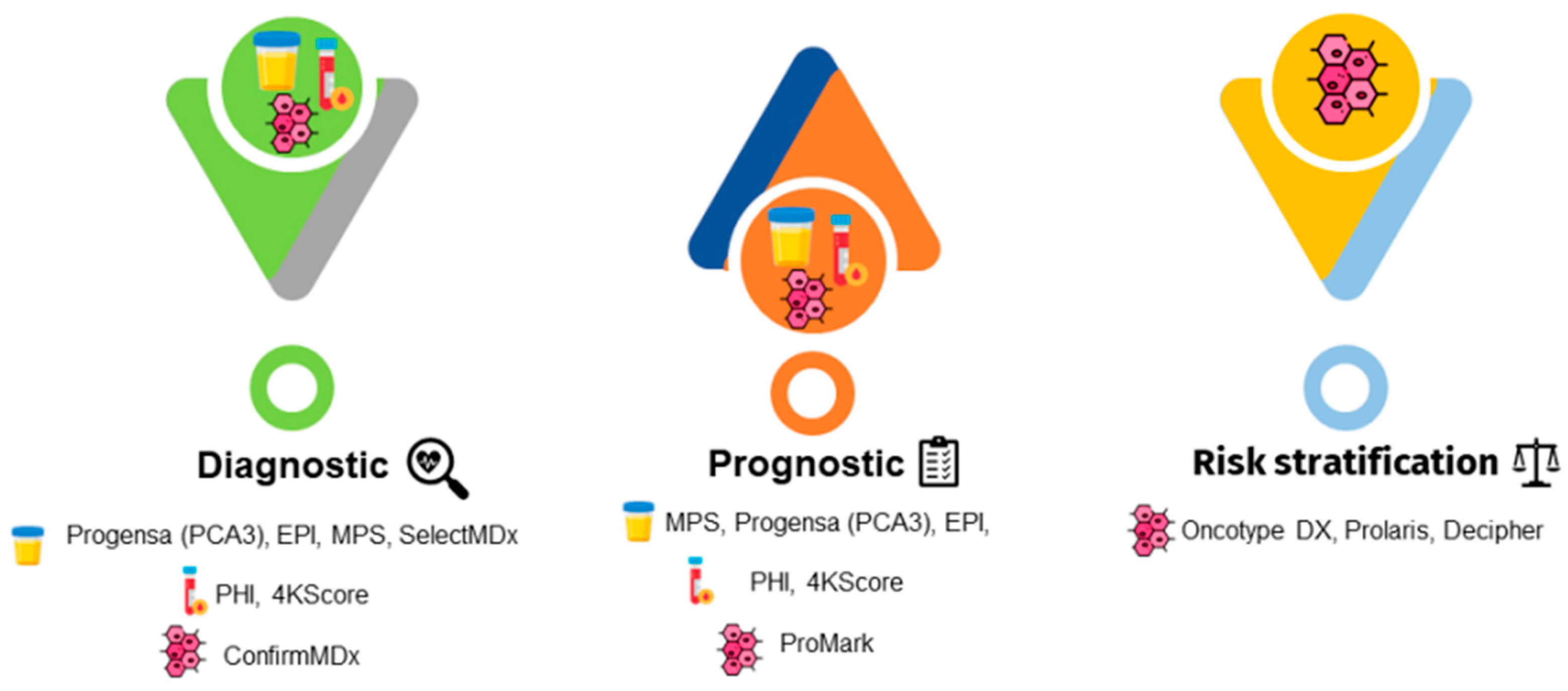
Figure 2. Emergent prostate cancer biomarkers for urine, serum, and tissue samples have been developed and can be used for diagnostic, prognostic, or risk stratification purposes. Legend: EPI, ExoDx Prostate IntelliScore; MPS, MyProstateScore; PCA3, prostate cancer gene 3; PHI, Prostate Health Index; 4KScore, Four-Kallikrein test.
Biomarkers such as the lncRNA PCA3 and TMPRSS2-ERG fusion gene have shown increased sensitivity and specificity (Table 1), potentially reducing PCa overdiagnosis. The prostate cancer gene 3 (PCA3) assay detects long non-coding RNA in urine samples. This test was approved by the Food and Drug Administration (FDA) in 2012; it calculates the ratio of PCA3 messenger RNAs (mRNAs) versus PSA mRNA in the first urine post-DRE and is approved for patients with a previously negative biopsy [7][10][32][33]. The Prostate Health Index (PHI) test is an algorithm approved by the FDA that includes total PSA, free PSA, and p2PSA isoform ([-2] proPSA). PHI calculates PCa probability and is recommended for men with PSA levels between 2 and 10 ng/mL and no abnormalities in their DRE. This blood test is also able to assess the likelihood of PCa progression during active surveillance, being used to monitor patients [34]. The Four-Kallikrein (4KScore) test is a diagnostic algorithm that combines four kallikreins in blood plasma, namely human kallikrein 2 (hK2), total PSA, free PSA, intact PSA, in addition to the patient’s clinical information (age, DRE results, and prostate biopsy history). This test assesses the probability of high-grade PCa and is recommended for patients undergoing initial and repeated biopsy. Moreover, the 4KScore also predicts the risk of occurrence and development of aggressive PCa [34]. The ExoDx Prostate IntelliScore (EPI) is a pre-biopsy RNA-based assay that uses the expression of PCA3, ERG, and SPDEF, isolated from urinary exosomes to predict the probability of high-grade PCa (Gleason score ≥ 7) on diagnostic biopsy. This is the only test that is not based on any other parameters related to PSA or a PSA derivative in the test algorithm to calculate the result, but clinicians can use it in conjugation with other clinical variables [30][34]. The SelectMDx and MyProstateScore (MPS) tests are based on the combination of multiple gene analyses. SelectMDx is a non-invasive test that measures mRNA transcripts from the genes HOXC6 and DLX1 in urine samples post-DRE and relates them to clinical risk factors such as age, family history, and PSA levels. This test is used to evaluate the presence of any PCa during biopsy and the risk of high-grade PCa. It also avoids unnecessary biopsies in the case of low-risk PCa [27]. Serum and urine biomarkers are used for consideration of initial biopsy, while tissue biomarkers are used to confirm test results. Tissue biomarkers tests have been developed to aid the clinical practice to decide what kind of therapy should be applied for different PCa diagnoses. ConfirmMDx is based on pronounced epigenetic changes that are indicative of the presence of cancer in the benign prostate tissue that is near the focus of PCa. This test determines the level of methylation of the promoter regions of the genes GSTP1, APC, and RASSF1 in benign prostate tissue, identifying high-grade PCa in patients with negative biopsies [27]. Prolaris is a prognostic test that measures tumor biology to improve the accuracy of risk stratification in men with localized PCa. This test combines the RNA expression levels of 31 genes involved in cell-cycle progression and 15 housekeeping genes to generate a Prolaris Score. Prolaris can be used to guide patient selection for active surveillance or definitive treatment [34][35][36]. The ProMark test is a protein-based prognostic assay of eight protein markers (DERL1, CUL2, SMAD4, PDSS2, HSPA9, FUS, phosphorylated S6, and YBOX1) that predicts the aggressiveness of cancer in patients with Gleason scores of 3 + 3 and 3 + 4. Moreover, ProMark predicts adverse pathology during radical prostatectomy and predicts if the tumor can be managed with or without aggressive treatment [27]. The Decipher is a genomic classifier prediction model for metastasis that measures the levels of RNA expression of 22 different genes on post-prostatectomy tissue samples. This test calculates the likelihood of clinical metastases within 5 years of prostatectomy in men with adverse pathological features. It could be a useful tool for diagnosis and local therapy planning for new PCa patients [31][34].
Despite the recent progress in the discovery of new biomarkers, gene mutations, and genomic signatures, some challenging obstacles must be overcome to develop effective biomarkers. These limitations include tumor heterogeneity, tumor–host interplay, complexity, multiplicity, and redundancy of tumor-cell signaling networks, involving genetic, epigenetic, and microenvironmental effects [7]. Additionally, the technologies associated with these approaches are often expensive, unavailable in many medical facilities, and time-consuming [10].
This entry is adapted from the peer-reviewed paper 10.3390/cancers14163982
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249.
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89.
- Beltran, H.; Demichelis, F. Intrapatient heterogeneity in prostate cancer. Nat. Rev. Urol. 2015, 12, 430–431.
- Dudka, I.; Thysell, E.; Lundquist, K.; Antti, H.; Iglesias-Gato, D.; Flores-Morales, A.; Bergh, A.; Wikström, P.; Gröbner, G. Comprehensive metabolomics analysis of prostate cancer tissue in relation to tumor aggressiveness and TMPRSS2-ERG fusion status. BMC Cancer 2020, 20, 437.
- Ostrander, E.A.; Johannesson, B. Prostate cancer susceptibility loci: Finding the genes. Adv. Exp. Med. Biol. 2008, 617, 179–190.
- Kgatle, M.M.; Kalla, A.A.; Islam, M.M.; Sathekge, M.; Moorad, R. Prostate Cancer: Epigenetic Alterations, Risk Factors, and Therapy. Prostate Cancer 2016, 2016, 1–11.
- Salciccia, S.; Capriotti, A.L.; Lagana, A.; Fais, S.; Logozzi, M.; De Berardinis, E.; Busetto, G.M.; Di Pierro, G.B.; Ricciuti, G.P.; Del Giudice, F.; et al. Biomarkers in Prostate Cancer Diagnosis: From Current Knowledge to the Role of Metabolomics and Exosomes. Int. J. Mol. Sci. 2021, 22, 4367.
- Culp, M.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52.
- Lee, S.; Ku, J.Y.; Kang, B.J.; Kim, K.H.; Ha, H.K.; Kim, S. A Unique Urinary Metabolic Feature for the Determination of Bladder Cancer, Prostate Cancer, and Renal Cell Carcinoma. Metabolites 2021, 11, 591.
- Lima, A.R.; Pinto, J.; Amaro, F.; Bastos, M.L.; Carvalho, M.; Guedes de Pinho, P. Advances and Perspectives in Prostate Cancer Biomarker Discovery in the Last 5 Years through Tissue and Urine Metabolomics. Metabolites 2021, 11, 181.
- Louie, K.S.; Seigneurin, A.; Cathcart, P.; Sasieni, P. Do prostate cancer risk models improve the predictive accuracy of PSA screening? A meta-analysis. Ann. Oncol. 2015, 26, 848–864.
- Das, C.J.; Razik, A.; Sharma, S.; Verma, S. Prostate biopsy: When and how to perform. Clin. Radiol. 2019, 74, 853–864.
- Spur, E.M.; Decelle, E.A.; Cheng, L.L. Metabolomic imaging of prostate cancer with magnetic resonance spectroscopy and mass spectrometry. Eur. J. Nucl. Med. Mol. Imaging 2013, 40 (Suppl. S1), S60–S71.
- Morrison, G.J.; Goldkorn, A. Development and Application of Liquid Biopsies in Metastatic Prostate Cancer. Curr. Oncol. Rep. 2018, 20, 35.
- Kretschmer, A.; Tilki, D. Biomarkers in prostate cancer–Current clinical utility and future perspectives. Crit. Rev. Oncol. Hematol. 2017, 120, 180–193.
- Xu, Y.-H.; Deng, J.-L.; Wang, G.; Zhu, Y.-S. Long non-coding RNAs in prostate cancer: Functional roles and clinical implications. Cancer Lett. 2019, 464, 37–55.
- Matuszczak, M.; Schalken, J.A.; Salagierski, M. Prostate Cancer Liquid Biopsy Biomarkers’ Clinical Utility in Diagnosis and Prognosis. Cancers 2021, 13, 3373.
- Oto, J.; Fernández-Pardo, Á.; Royo, M.; Hervás, D.; Martos, L.; Vera-Donoso, C.D.; Martínez, M.; Heeb, M.J.; España, F.; Medina, P.; et al. A predictive model for prostate cancer incorporating PSA molecular forms and age. Sci. Rep. 2020, 10, 2463.
- Auprich, M.; Bjartell, A.; Chun, F.K.; de la Taille, A.; Freedland, S.J.; Haese, A.; Schalken, J.; Stenzl, A.; Tombal, B.; van der Poel, H. Contemporary role of prostate cancer antigen 3 in the management of prostate cancer. Eur. Urol. 2011, 60, 1045–1054.
- Parekh, D.J.; Punnen, S.; Sjoberg, D.D.; Asroff, S.W.; Bailen, J.L.; Cochran, J.S.; Concepcion, R.; David, R.D.; Deck, K.B.; Dumbadze, I.; et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur. Urol. 2015, 68, 464–470.
- Nordström, T.; Vickers, A.; Assel, M.; Lilja, H.; Grönberg, H.; Eklund, M. Comparison Between the Four-kallikrein Panel and Prostate Health Index for Predicting Prostate Cancer. Eur. Urol. 2015, 68, 139–146.
- Zappala, S.M.; Scardino, P.T.; Okrongly, D.; Linder, V.; Dong, Y. Clinical performance of the 4Kscore Test to predict high-grade prostate cancer at biopsy: A meta-analysis of us and European clinical validation study results. Rev. Urol. 2017, 19, 149–155.
- White, J.; Shenoy, B.V.; Tutrone, R.F.; Karsh, L.I.; Saltzstein, D.R.; Harmon, W.J.; Broyles, D.L.; Roddy, T.E.; Lofaro, L.R.; Paoli, C.J.; et al. Clinical utility of the Prostate Health Index (phi) for biopsy decision management in a large group urology practice setting. Prostate Cancer Prostatic Dis. 2018, 21, 78–84.
- Saidi, S.; Al Riyami, N.; Marhoon, M.; Saraf, M.; Busaidi, S.; Mula-Abed, W.-A.; Bayoumi, R. Validity of Prostate Health Index and Percentage of Pro-Prostate-Specific Antigen as Novel Biomarkers in the Diagnosis of Prostate Cancer: Omani Tertiary Hospitals Experience. Oman Med. J. 2017, 32, 275–283.
- Rodríguez, S.V.M.; García-Perdomo, H.A. Diagnostic accuracy of prostate cancer antigen 3 (PCA3) prior to first prostate biopsy: A systematic review and meta-analysis. Can. Urol. Assoc. J. 2020, 14, E214–E219.
- Nicholson, A.; Mahon, J.; Boland, A.; Beale, S.; Dwan, K.; Fleeman, N.; Hockenhull, J.; Dundar, Y. The clinical effectiveness and cost-effectiveness of the PROGENSA® prostate cancer antigen 3 assay and the Prostate Health Index in the diagnosis of prostate cancer: A systematic review and economic evaluation. Health Technol. Assess. 2015, 19, 1–191.
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur. Urol. 2016, 70, 740–748.
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53.
- BridgeSpan. Gene-Based Tests for Screening, Detection, and/or Management of Prostate Cancer. In Genetic Testing, Policy No. 17; BridgeSpan: Boston, MA, USA, 2020.
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentinck, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889.
- Ross, A.E.; Feng, F.Y.; Ghadessi, M.; Echo, N.; Crisan, A.; Buerki, C.; Sundi, D.; Mitra, A.P.; Vergara, I.A.; Thompson, D.J.; et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 2014, 17, 64–69.
- Narayan, V.M.; Konety, B.R.; Warlick, C. Novel biomarkers for prostate cancer: An evidence-based review for use in clinical practice. Int. J. Urol. 2017, 24, 352–360.
- Chistiakov, D.A.; Myasoedova, V.A.; Grechko, A.V.; Melnichenko, A.A.; Orekhov, A.N. New biomarkers for diagnosis and prognosis of localized prostate cancer. Semin. Cancer Biol. 2018, 52, 9–16.
- Porzycki, P.; Ciszkowicz, E. Modern biomarkers in prostate cancer diagnosis. Cent. Eur. J. Urol. 2020, 73, 300–306.
- Crawford, E.D.; Ventii, K.; Shore, N.D. New biomarkers in prostate cancer. Oncology 2014, 28, 135–142.
- Clinton, T.N.; Bagrodia, A.; Lotan, Y.; Margulis, V.; Raj, G.V.; Woldu, S.L. Tissue-based biomarkers in prostate cancer. Expert Rev. Precis. Med. Drug Dev. 2017, 2, 249–260.
This entry is offline, you can click here to edit this entry!
