Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Inflammatory bowel disease (IBD), classified primarily between Crohn’s disease and ulcerative colitis, is a collection of chronic gastrointestinal inflammatory conditions that cause multiple complications because of systemic alterations in the immune response. IBD is currently diagnosed through a multitude of different assessments including clinical history, radiology, endoscopy, colonoscopy, and histology. Diagnostic challenges remain in differentiating between UC and CD when lesions are solely limited to the colon and in differentiating between IBD and irritable bowel syndrome (IBS). Endoscopy represents the main method of differentiation between organic IBD, and more functional IBS disorder though inflammatory markers such as TNF-α and calprotectin has also been used. miRNAs are found to be stable in peripheral blood, saliva, and feces and have been suggested as diagnostic biomarkers of IBD. There is also research indicating that miRNAs can serve as sensitive and specific biomarkers for disease onset, prognosis, and remission.
- Crohn’s disease
- ulcerative colitis
- non-coding RNA
1. Differentiating UC and CD with miRNAs
While distinguishing between UC and CD remains a diagnostic challenge, newer biomarkers such as human alpha defensin 5 and miRNAs may aid in diagnosis, especially in histologically indeterminate scenarios [1]. Forming a miRNA panel may help serve as a potential tool to distinguish between CD and UC and evaluate complications of IBD, as discussed. In addition, utilizing differential miRNAs for diagnosis is much less invasive than classic endoscopy, and yet as specific [2][3]. Based on the literature, the researchers created Venn diagrams showing common differentially dysregulated miRNAs in the ileal/colonic tissue (Figure 1A), peripheral blood (Figure 1B), feces (Figure 1C), and saliva (Figure 1D) of patients with UC or CD. Albeit that further validation is needed, different miRNA panels could be established and utilized for differential diagnosis of UC and CD.
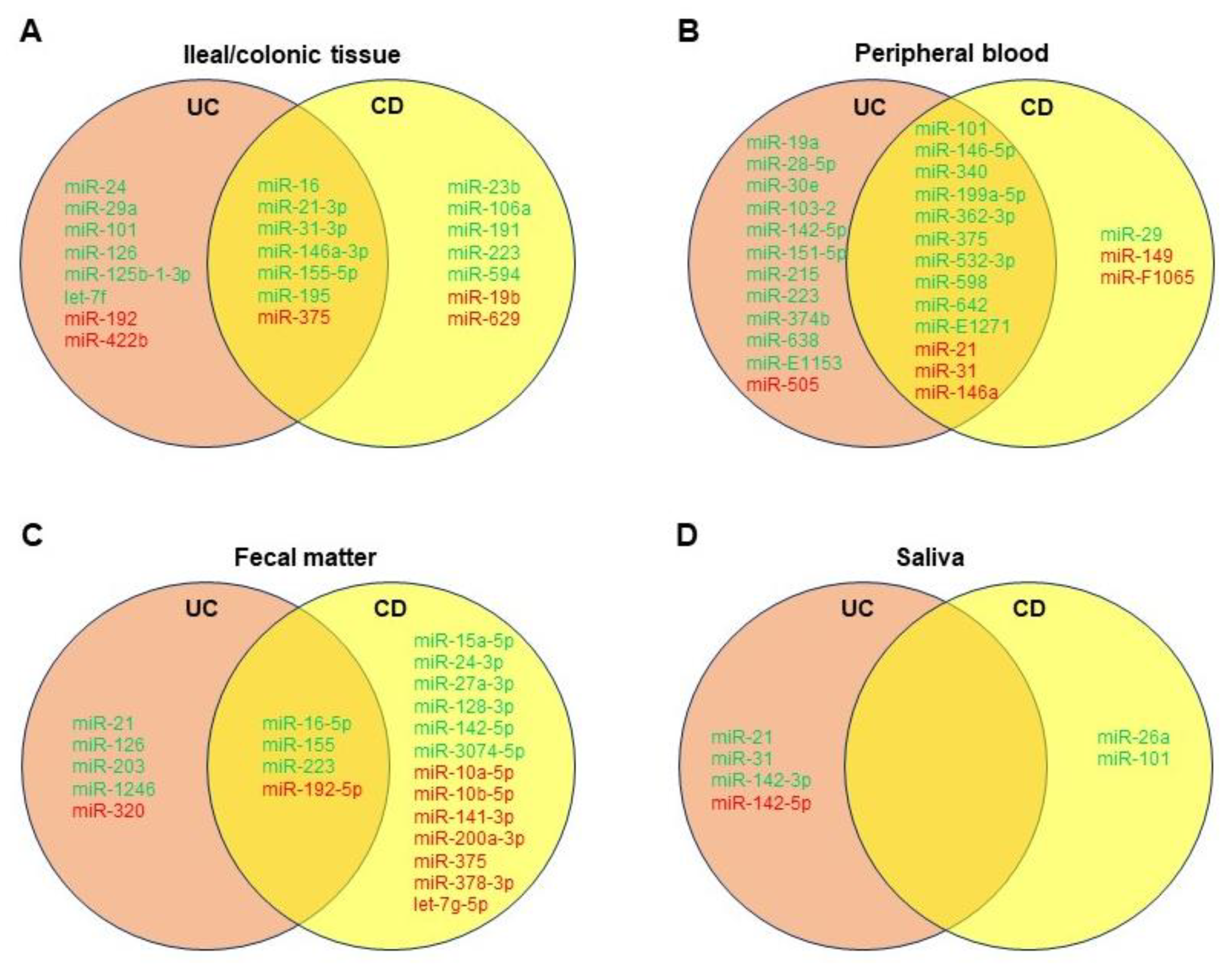
Figure 1. Venn diagrams showing significantly dysregulated miRNAs in ileal/colonic tissue (A), peripheral blood (B), fecal matter (C), and saliva (D) of patients with ulcerative colitis (UC) or Crohn’s disease (CD). Green denotes upregulated miRNAs, and red denotes downregulated miRNAs compared to healthy controls.
One study from Wu et al. noted eight confirmed miRNAs in peripheral blood that were distinguishable between active UC and active CD: miRs-28-5p, -103-2, -149, -151-5p, -340, -532-3p, and miR-plus-E1153 were all significantly elevated in active UC when compared to active CD, and miR-505 was significantly decreased in active UC versus active CD [3]. The study also noted these miRNA “signatures” were more homogenous in the peripheral blood samples of active disease patients as opposed to inactive remission patients. Another study determined after screening assays that miRs-598 and -642 were significantly upregulated in the UC patients’ plasma in comparison to CD patients’, though both were significantly elevated in UC and CD compared to healthy controls [4]. Furthermore, miRs-16, -21, and, in particular, -223 were more prominently increased in active CD sera versus that of active UC [5].
In fecal matter, miRs-223 and -1246 were significantly upregulated in UC individuals with fecal calprotectin > 250 mg/kg compared to CD individuals with fecal calprotectin < 250 mg/kg [6]. Studies from Ahmed et al. and Wohnhaas et al. seem to suggest upregulation of miRs-21, -126, and -203 and downregulation of miR-320 may distinguish UC from CD [7][8]. In general, it seems proinflammatory miRNA expression is higher-fold in feces of UC patients compared to CD patients, though this depends on other variables as well, including disease activity [5]. Salivary miRNAs are still quite novel, it seems miRs-26a and -101 may be more prominent in CD patients [2].
Even with colonic tissue biopsies, a more specific and complementary miRNA panel may better support histology if colonoscopy is to be undertaken. For example, although miR-29a is significantly upregulated in UC colon samples, miR-29 family members in CD colon tissue are not, with downregulation even occurring along strictures via TGF-β-mediated fibrosis [9]. Furthermore, miR-125b-1-3p was determined by Schaefer et al. as only significantly upregulated in UC colon tissue [2]. Other miRNAs to consider may include miRs-31, -106a, -146a, -192, and -375, though further research with larger sample sizes with attention toward disease activity may improve the knowledge of various miRNAs involved in either UC or CD.
One diagnostic difficulty in IBD is the differentiation between clinical and endoscopic remission, where clinical remission is the absence of symptoms, but endoscopic remission is the absence of detectable mucosal lesions and is associated with better clinical prognosis [10]. miR-320a was found to be increased in the peripheral blood of CD patients with active disease flares compared to those with quiescent disease; was directly correlated with disease severity; was associated with endoscopic disease activity in the setting of mild or absent clinical symptoms; was increased in CD patients with extensive intestinal involvement; and was elevated compared to patients with C. difficile-associated colitis [10][11]. While miR-320a is a candidate for being a diagnostic biomarker for detecting differentiation of active colitis, Cordes et al. showed that it does not correlate with histological colitis activity, and larger cohort studies are needed to fully assess the diagnostic value [12]. For serum miR-146b-5p, CD and UC patients had levels 2.87-fold higher and 2.72-fold higher than age- and gender-matched healthy controls. Chen et al. found that miR-146-5p had potential diagnostic value as its expression had similar sensitivity to CRP, a marker that is associated with disease activity in UC/CD, and displayed increased specificity compared to CRP (92.31% vs. 46.15%).
2. Differentiating IBS and IBD with miRNAs
miRNAs may help differentiate IBS from IBD and its complications. One study noted significant upregulation of miRs-23a, -375, and -422b in IBS colonic tissue compared to healthy controls [13]. Zhou et al. found upregulated miRs-29a and -29b in the small intestinal and colonic tissue of IBS-D patients [14]. In colonic mucosa, miRs-219a-5p and -338-3p were found downregulated in IBS patients [15]. miR-375 may also be useful for IBS/IBD differentiability exhibiting downregulation in IBD colonic tissue, in contrast to IBS upregulation. For CD specifically, miR-29b may distinguish CD versus IBS with downregulation and upregulation, respectively.
In serum studies, proinflammatory miRs-23a and -181b are upregulated in IBS patients compared to healthy controls [16]. Moreover, miRs-150 and -342-3p were also found upregulated in IBS patients [17]. Conversely, serum miR-199b levels were downregulated [18]. Another study identified miRs-21 and -92a upregulated in UC compared to IBS patients [19]. Just like miRNAs can distinguish UC and CD, more research may help solidify differential miRNAs in IBS patients with consideration for specific disease variation amongst IBS-C, IBS-D, and IBS-M.
3. Challenges and Future Indications for miRNA-Based Diagnostics
Some challenges that miRNAs face as a diagnostic profiling tool is the challenge of normalizing peripheral fluid data to obtain standard cut-off values, low sensitivities and specificities in current detection methods, and creating point-of-care assays to decrease diagnostic latency [20][21][22]. While miRNAs may prove to be useful diagnostic prognosticators, more information is needed with regard to tissue/sera normalization across a cohort with a wide variety of patient backgrounds to increase validity. Furthermore, because of the multiplicity of roles that a single miRNA is involved in, it may be useful to try using a panel-based approach, as discussed above, for future research and diagnostic purposes. Current detection methods of miRNAs include traditional methods such as Northern blotting, microarrays, and RT-qPCR, while newer detection methods include nanomaterial-based miRNA detection, nucleic acid amplification techniques, rolling circle amplification, fluorescent in situ hybridization, strand displacement amplification, loop-mediated isothermal amplification, and enzyme-free amplification [21]. While traditional RNA detection methods are predominantly employed, these methods suffer from limitations such as low specificity and sensitivity and applications requiring technical expertise and can be time consuming. Newer detection techniques display increased sensitivity and specificity and increased discriminatory ability and can potentially be more cost-effective. These technologies may play a role in point-of-care settings as microfluidic chip- and electrochemical-based systems augment these newer detection routes to become more portable than current diagnostics modalities [22]. However, these newer techniques still display limitations such as cost and complexity, as well as require further sensitivity and specificity validation. The future of miRNA detection will require a widespread validation of newer techniques, as well as the employment of a combination of existing techniques to utilize miRNA diagnostics in a cost-effective, widespread manner.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23158751
References
- Williams, A.D.; Korolkova, O.Y.; Sakwe, A.M.; Geiger, T.M.; James, S.D.; Muldoon, R.L.; Herline, A.J.; Goodwin, J.S.; Izban, M.G.; Washington, M.K.; et al. Human alpha defensin 5 is a candidate biomarker to delineate inflammatory bowel disease. PLoS ONE 2017, 12, e0179710.
- Schaefer, J.S.; Attumi, T.; Opekun, A.R.; Abraham, B.; Hou, J.; Shelby, H.; Graham, D.Y.; Streckfus, C.; Klein, J.R. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015, 16, 5.
- Wu, F.; Guo, N.J.; Tian, H.; Marohn, M.; Gearhart, S.; Bayless, T.M.; Brant, S.R.; Kwon, J.H. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm. Bowel Dis. 2011, 17, 241–250.
- Netz, U.; Carter, J.; Eichenberger, M.R.; Feagins, K.; Galbraith, N.J.; Dryden, G.W.; Pan, J.; Rai, S.N.; Galandiuk, S. Plasma microRNA Profile Differentiates Crohn’s Colitis From Ulcerative Colitis. Inflamm. Bowel Dis. 2017, 24, 159–165.
- Schonauen, K.; Le, N.; von Arnim, U.; Schulz, C.; Malfertheiner, P.; Link, A. Circulating and Fecal microRNAs as Biomarkers for Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2018, 24, 1547–1557.
- Verdier, J.; Breunig, I.R.; Ohse, M.C.; Roubrocks, S.; Kleinfeld, S.; Roy, S.; Streetz, K.; Trautwein, C.; Roderburg, C.; Sellge, G. Faecal Micro-RNAs in Inflammatory Bowel Diseases. J. Crohns Colitis 2020, 14, 110–117.
- Ahmed, F.E.; Jeffries, C.D.; Vos, P.W.; Flake, G.; Nuovo, G.J.; Sinar, D.R.; Naziri, W.; Marcuard, S.P. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genom. Proteom. 2009, 6, 281–295.
- Wohnhaas, C.T.; Schmid, R.; Rolser, M.; Kaaru, E.; Langgartner, D.; Rieber, K.; Strobel, B.; Eisele, C.; Wiech, F.; Jakob, I.; et al. Fecal MicroRNAs Show Promise as Noninvasive Crohn’s Disease Biomarkers. Crohns Colitis 360 2020, 2, otaa003.
- Nijhuis, A.; Biancheri, P.; Lewis, A.; Bishop, C.L.; Giuffrida, P.; Chan, C.; Feakins, R.; Poulsom, R.; Di Sabatino, A.; Corazza, G.R.; et al. In Crohn’s disease fibrosis-reduced expression of the miR-29 family enhances collagen expression in intestinal fibroblasts. Clin. Sci. (Lond.) 2014, 127, 341–350.
- Cordes, F.; Demmig, C.; Bokemeyer, A.; Brückner, M.; Lenze, F.; Lenz, P.; Nowacki, T.; Tepasse, P.; Schmidt, H.H.; Schmidt, M.A.; et al. MicroRNA-320a Monitors Intestinal Disease Activity in Patients With Inflammatory Bowel Disease. Clin. Transl. Gastroenterol. 2020, 11, e00134.
- Cordes, F.; Brückner, M.; Lenz, P.; Veltman, K.; Glauben, R.; Siegmund, B.; Hengst, K.; Schmidt, M.A.; Cichon, C.; Bettenworth, D. MicroRNA-320a Strengthens Intestinal Barrier Function and Follows the Course of Experimental Colitis. Inflamm. Bowel Dis. 2016, 22, 2341–2355.
- Chen, P.; Li, Y.; Li, L.; Yu, Q.; Chao, K.; Zhou, G.; Qiu, Y.; Feng, R.; Huang, S.; He, Y.; et al. Circulating microRNA146b-5p is superior to C-reactive protein as a novel biomarker for monitoring inflammatory bowel disease. Aliment. Pharmacol. Ther. 2019, 49, 733–743.
- Wu, F.; Zikusoka, M.; Trindade, A.; Dassopoulos, T.; Harris, M.L.; Bayless, T.M.; Brant, S.R.; Chakravarti, S.; Kwon, J.H. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 2008, 135, 1624–1635.e24.
- Zhou, Q.; Costinean, S.; Croce, C.M.; Brasier, A.R.; Merwat, S.; Larson, S.A.; Basra, S.; Verne, G.N. MicroRNA 29 targets nuclear factor-κB-repressing factor and Claudin 1 to increase intestinal permeability. Gastroenterology 2015, 148, 158–169.e8.
- Mahurkar-Joshi, S.; Rankin, C.R.; Videlock, E.J.; Soroosh, A.; Verma, A.; Khandadash, A.; Iliopoulos, D.; Pothoulakis, C.; Mayer, E.A.; Chang, L. The Colonic Mucosal MicroRNAs, MicroRNA-219a-5p, and MicroRNA-338-3p Are Downregulated in Irritable Bowel Syndrome and Are Associated With Barrier Function and MAPK Signaling. Gastroenterology 2021, 160, 2409–2422.e19.
- Chira, A.; Muresan, M.S.; Braicu, C.; Budisan, L.; Raduly, L.; Chira, R.I.; Dumitrascu, D.L.; Berindan-Neagoe, I. Serum patterns of mir-23a and mir-181b in irritable bowel syndrome and colorectal cancer—A pilot study. Bosn. J. Basic Med. Sci. 2020, 20, 254–261.
- Fourie, N.H.; Peace, R.M.; Abey, S.K.; Sherwin, L.B.; Rahim-Williams, B.; Smyser, P.A.; Wiley, J.W.; Henderson, W.A. Elevated circulating miR-150 and miR-342-3p in patients with irritable bowel syndrome. Exp. Mol. Pathol. 2014, 96, 422–425.
- Mansour, M.A.; Sabbah, N.A.; Mansour, S.A.; Ibrahim, A.M. MicroRNA-199b expression level and coliform count in irritable bowel syndrome. IUBMB Life 2016, 68, 335–342.
- Ahmed Hassan, E.; El-Din Abd El-Rehim, A.S.; Mohammed Kholef, E.F.; Abd-Elgwad Elsewify, W. Potential role of plasma miR-21 and miR-92a in distinguishing between irritable bowel syndrome, ulcerative colitis, and colorectal cancer. Gastroenterol. Hepatol. Bed Bench 2020, 13, 147–154.
- James, J.P.; Riis, L.B.; Malham, M.; Høgdall, E.; Langholz, E.; Nielsen, B.S. MicroRNA Biomarkers in IBD-Differential Diagnosis and Prediction of Colitis-Associated Cancer. Int. J. Mol. Sci. 2020, 21, 7893.
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226.
- Dave, V.P.; Ngo, T.A.; Pernestig, A.K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Invest. 2019, 99, 452–469.
This entry is offline, you can click here to edit this entry!
