Developing varieties with diverse features that satisfy varied commercial needs, improving overall fruit quality, and quickly releasing them, are prerequisites in citrus breeding. However, these three goals require trade-offs in conventional breeding, even with the application of the marker-assisted selection technique. Conventional breeding cannot achieve these three goals simultaneously and it has been regarded as a breeding trilemma. Integrating a genomics-assisted breeding (GAB) approach that relies on quantitative trait locus detection by genome-wide association study and genome-wide prediction of a trait by genomic selection using enriched marker genotypes enhances breeding efficiency and contributes to eliminating the trilemma. Besides these efforts, the analysis of the genealogy of indigenous citrus varieties revealed that many high-quality indigenous varieties were selected within a few generations. It suggested that selecting a new premium quality hybrid is possible by integrating it with the GAB technique and helps avoid the trilemma. Researchers describe an ongoing comprehensive approach for integrating genomic-assisted breeding (GAB) with citrus genealogy on citrus breeding, called Citrus Breeding 2.0. This method can develop new cultivars with premium quality in a short period.
- citrus breeding
- marker-assisted selection
- genomics-assisted breeding
- genome-wide association study
- genomic selection
- genealogy
1. Introduction
2. Citrus Crossbreeding Effort Aims to Improve Fruit Quality
2.1. Past Efforts of the Citrus Breeding Program in NARO
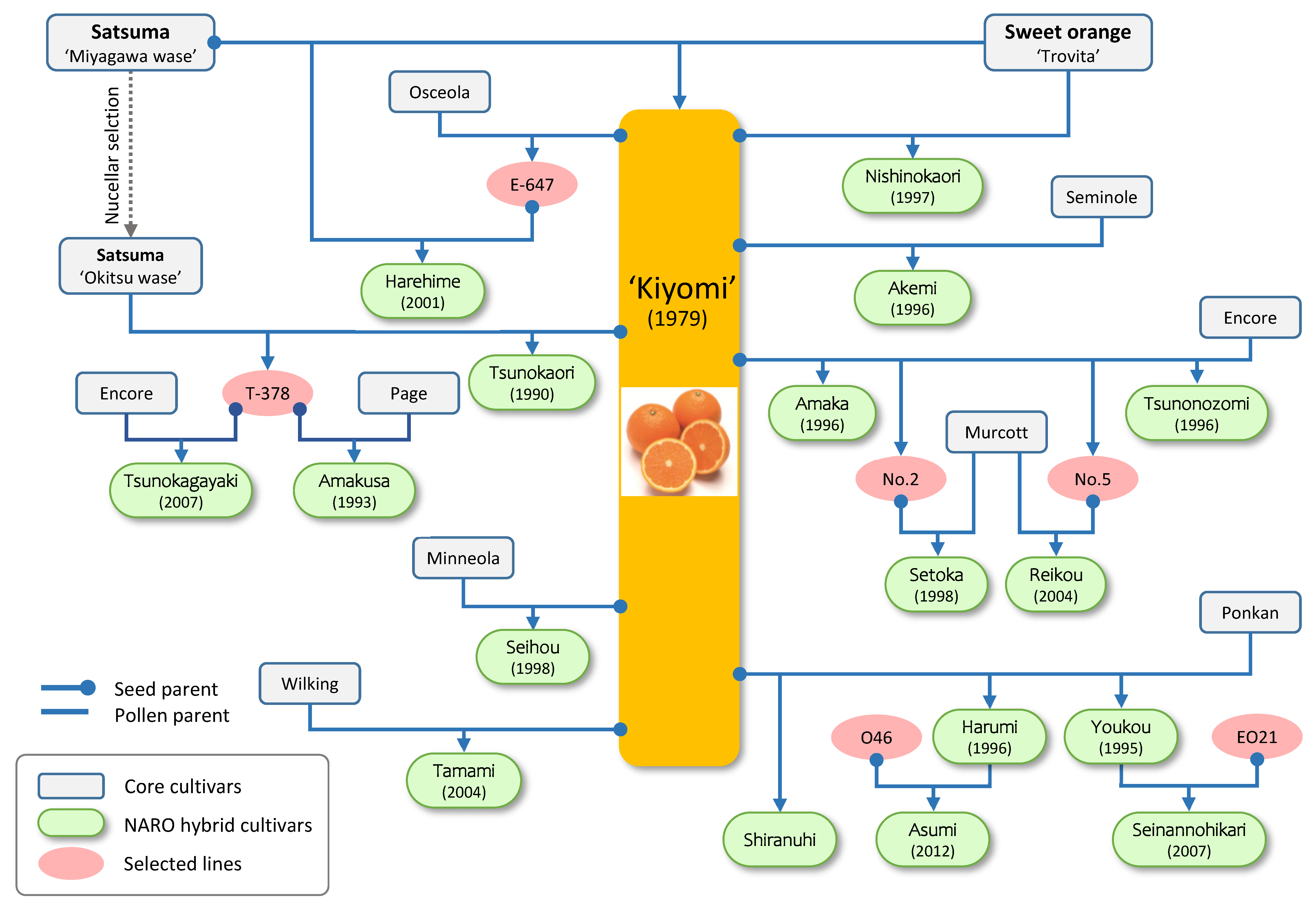
| Cultivar | Year of Release | YTD | Harvest | Fruit Size (g) |
Peelability | Seeds | Features |
|---|---|---|---|---|---|---|---|
| Kiyomi | 1979 | 30 | Late March | ~200 | Moderate | SL | A tangor (mandarin x orange). Orange-like aroma and juicy |
| Seihou | 1988 | 17 | Late January | ~200 | Hard | SL | Large fruit with orange-like good flavor and soft flesh |
| Tshunokaori | 1990 | 18 | March–April | ~160 | Easy | SL | High SSC (>13), and no rind-puffing |
| Amakusa | 1993 | 11 | August–January | ~200 | Hard | -- | High SSC (>12), and less alternate bearing |
| Youkou | 1995 | 23 | January–February | 250–300 | Easy | SL | Large fruit with good flavor and soft flesh |
| Harumi | 1996 | 17 | January | 180–200 | Easy | SL | Large fruit with good flavor and soft flesh |
| Akemi | 1996 | 21 | March | 160–180 | Moderate | SL | High SSC (>12), grenadine rind, and soft flesh |
| Amaka | 1996 | 22 | August–January | 200–250 | Easy | SL | Soft fruit with good orange-like aroma |
| Nishinokaori | 1997 | 31 | August–January | 100–180 | Easy | SL | High SSC (>12), orange-like flavor, and soft flesh |
| Setoka | 1998 | 14 | February | 200–280 | Easy | SL | Large fruit with good flavor, high SSC (>12), and soft flesh |
| Harehime | 2001 | 11 | August | 180 | Easy | SL | Early type with good flavor, juicy, and soft flesh |
| Tamami | 2004 | 24 | January | 150 | Easy | -- | High SSC (>12) and good orange-like flavor |
| Reikou | 2004 | 20 | January | 210 | Moderate | SL | High SSC (>12), good flavor, and red-skin |
| Tsunokagayaki | 2008 | 24 | January–February | 180–250 | Easy | SL | High SSC (>13), soft fruit, BCP rich, and suitable for greenhouse production |
| Tsunonozomi | 2011 | 37 | August | 190 | Easy | -- | Early type with good flavor, high SSC (>12), juicy, and less rind puffing |
| Asumi | 2013 | 21 | February | 150 | Moderate | SL | Very high SSC (>15), orange-like flavor, and BCP rich |
| Shiranuhi * | February–March | 230 | Easy | SL | High SSC (>14), soft fruit, good flavor, and unique fruit shape |
2.2. Constraints of Conventional Crossbreeding
3. Suggestions from the Citrus Genealogy for Breeding
3.1. Genomic Breeding Aims to Avoid the Breeding Trilemma
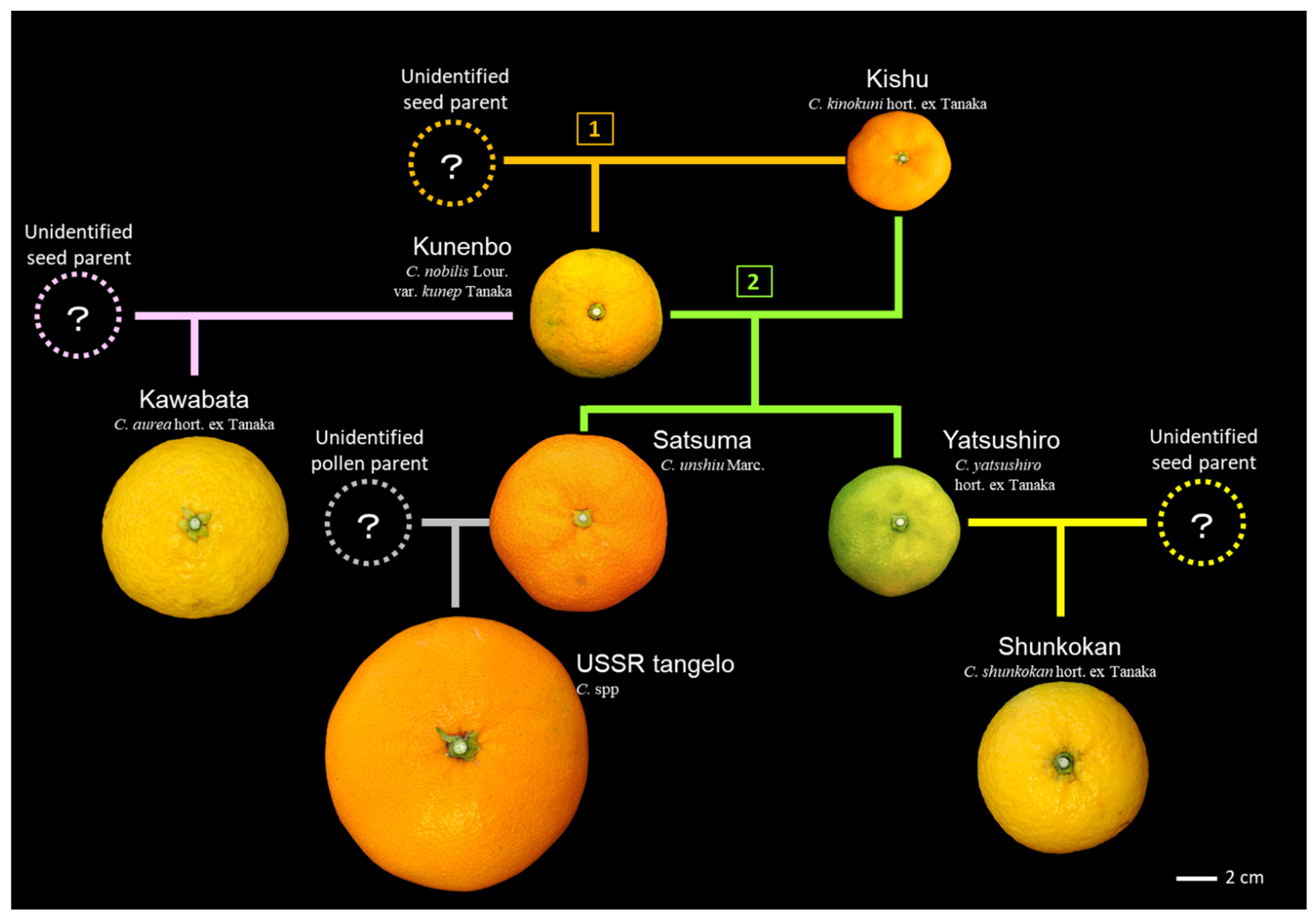
3.2. Lessons from the Revealed Genealogy of Citrus Cultivars
3.3. Hybrids Selected from Kishu and Kunenbo
This entry is adapted from the peer-reviewed paper 10.3390/horticulturae8060559
References
- Webber, H.J. History and development of the citrus industry. In The Citrus Industry I.; Reuther, W., Webber, H.J., Batchelor, L.D., Eds.; University of California Press: Berkeley, CA, USA, 1967; pp. 1–39.
- Food and Agriculture Organization, Intergovernmental Group on Citrus Fruits. Citrus Fruit Statistics 2015; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016.
- Caruso, M.; Smith, M.W.; Froelicher, Y.; Russo, G.; Gmitter, F.G. Traditional breeding. In The Genus Citrus; Elsevier: Duxford, UK, 2020; pp. 129–148.
- Nishiura, M. Citrus breeding through nucellar seedling selection. JARQ Jpn. Agric. Res. Q. 1967, 2, 15–19.
- Lamo, K.; Ji Bhat, D.; Kour, K.; Singh Solanki, S.P. Mutation Studies in Fruit Crops: A Review. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3620–3633.
- Kamatyanatt, M.; Singh, S.K.; Sekhon, B.S. Mutation breeding in citrus—A review. Plant Cell Biotechnol. Mol. Biol. 2021, 22, 1–8.
- Roose, M.L.; Williams, T.E. Mutation breeding. In Citrus Genetics, Breeding and Biotechnology; Khan, I.A., Ed.; CABI, CAB International: Oxfordshire, UK, 2007; pp. 345–352. ISBN 978-0851990194.
- Cimen, B.; Yesiloglu, T.; Incesu, M.; Yilmaz, B. Studies on mutation breeding in citrus: Improving seedless types of ‘Kozan’ common orange by gamma irradiation. Sci. Hortic. 2021, 278, 109857.
- Iwamasa, M.; Yamaguchi, S.; Kuriyama, T.; Nakamuta, T.; Ehara, T.; Nito, N.; Katayama, Y. Occurrence of very early mutants from the wase (early ripening) satsumas and their characteristics. Bull. Fac. Agric. Saga Univ. 1984, 55, 99–107.
- Ollitrault, P.; Navarro, L. Citrus. In Fruit Breeding; Badenes, M.L., Byrne, D.H., Eds.; Springer: Boston, MA, USA, 2012; pp. 623–662. ISBN 9781441907639.
- Raveh, E.; Goldenberg, L.; Porat, R.; Carmi, N.; Gentile, A.; La Malfa, S. Conventional Breeding of Cultivated Citrus Varieties. In The Citrus Genome; Springer Nature: Cham, Switzerland, 2020; pp. 33–48.
- Shimizu, T.; Aka Kacar, Y.; Cristofani-Yaly, M.; Curtolo, M.; Machado, M.A. Markers, Maps, and Marker-Assisted Selection. In The Citrus Genome; Gentile, A., La Malfa, S.D.Z., Eds.; Springer: Cham, Switzerland, 2020; pp. 107–139. ISBN 978-3-030-10799-4.
- Shimizu, T. Citrus Breeding 2.0: A novel approach integrating deciphered parentage and genomics-assisted selection. JARQ Jpn. Agric. Res. Q. 2019, 53, 81–85.
- Nishiura, M. Citrus breeding and bud selection in Japan. Proc. Fla. State Hort. Soc. 1964, 77, 79–83.
- Nishiura, M.; Shichijo, T.; Ueno, I.; Iwamasa, M.; Kihara, T.; Yamada, Y.; Yoshida, T.; Iwasaki, T. New citrus cultivar “Kiyomi” tangor. Bull. Fruit Tree Res. Stn. Ser. B Okitsu 1983, 10, 1–9. (In Japanese)
- NARO: National Agriculture and Food Research Organization List of the Citrus Bred Varieties. Available online: https://www.naro.go.jp/laboratory/nifts/kih/hinshu/citrus_cat/index.html (accessed on 10 April 2022). (In Japanese).
- Shimizu, T. Genomic breeding. In The Genus Citrus; Talon, M., Gmitter, F., Caruso, M., Eds.; Elsevier: Duxford, UK, 2020; pp. 149–169. ISBN 978-0-12-812163-4.
- Iwata, H.; Minamikawa, M.F.; Kajiya-Kanegae, H.; Ishimori, M.; Hayashi, T. Genomics-assisted breeding in fruit trees. Breed. Sci. 2016, 66, 100–115.
- Minamikawa, M.F.; Nonaka, K.; Kaminuma, E.; Kajiya-Kanegae, H.; Onogi, A.; Goto, S.; Yoshioka, T.; Imai, A.; Hamada, H.; Hayashi, T.; et al. Genome-wide association study and genomic prediction in citrus: Potential of genomics-assisted breeding for fruit quality traits. Sci. Rep. 2017, 7, 4721.
- Combrink, N.K.; Labuschagne, M.T.; Bijzet, Z. Variation of fruit size and shape in Kiyomi tangor families. Sci. Hortic. 2013, 162, 357–364.
- Combrink, N.K.; Bijzet, Z.; Sippel, A.D.; Booyse, M.; Labuschagne, M.T. Genotypic variation of rind colour in citrus tangor Kiyomi families. Acta Hortic. 2015, 1065, 439–448.
- Kita, M.; Nesumi, H.; Kuniga, T.; Nakajima, N.; Yoshioka, T.; Ohta, S.; Takishita, F.; Nakano, M.; Ogawa, K.; Yoshida, T.; et al. New Citrus Cultivar ‘Aurastar’. J. NARO Res. Dev. 2021, 7, 1–8. (In Japanese)
- Yoshida, T.; Nesumi, H.; Nakajima, N.; Kuniga, T. New citrus parental lines “Kankitsu Chukanbohon Nou 7 Gou” and “Kankitsu Chukanbohon Nou 8 Gou” for breeding new cultivars with immunity to citrus tristeza virus. Jpn. J. Hortic. Sci. 2005, 74, 323. (In Japanese)
- Serra, O.; Donoso, J.M.; Picañol, R.; Batlle, I.; Howad, W.; Eduardo, I.; Arús, P. Marker-assisted introgression (MAI) of almond genes into the peach background: A fast method to mine and integrate novel variation from exotic sources in long intergeneration species. Tree Genet. Genomes 2016, 12, 96.
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316.
- Wu, G.A.; Sugimoto, C.; Kinjo, H.; Azama, C.; Mitsube, F.; Talon, M.; Gmitter, F.G.; Rokhsar, D.S. Diversification of mandarin citrus by hybrid speciation and apomixis. Nat. Commun. 2021, 12, 4377.
- Shimizu, T.; Kitajima, A.; Nonaka, K.; Yoshioka, T.; Ohta, S.; Goto, S.; Toyoda, A.; Fujiyama, A.; Mochizuki, T.; Nagasaki, H.; et al. Hybrid origins of citrus varieties inferred from DNA marker analysis of nuclear and organelle genomes. PLoS ONE 2016, 11, e0166969.
- Tanaka, T. Species Problem in Citrus: A Critical Study of Wild and Cultivated Units of Citrus, Based upon Field Studies in Their Native Homes (Revisio Aurantiacearum IX); Japanese Society for the Promotion of Science: Tokyo, Japan, 1954.
- Tanaka, T. A Revision of Osmocitrus, a section of the genus Citrus: Revisio Aurantiacearum XIII. Bull. Univ. Osaka Prefect. Ser. B Agric. Biol. 1960, 10, 9–13.
- Shimizu, T.; Kitajima, A.; Nonaka, K.; Yoshioka, T.; Ohta, S.; Goto, S.; Kaminuma, E.; Nakamura, Y. A model for the domestication and diversification processes of modern citrus cultivars in Japan. In Proceedings of the IV International Symposium on Citrus Biotechnology, Canelones, Uruguay, 16–18 April 2018; pp. 7–14.
- Shimizu, T.; Tanizawa, Y.; Mochizuki, T.; Nagasaki, H.; Yoshioka, T.; Toyoda, A.; Fujiyama, A.; Kaminuma, E.; Nakamura, Y. Draft sequencing of the heterozygous diploid genome of Satsuma (Citrus unshiu Marc.) using a hybrid assembly approach. Front. Genet. 2017, 8, 180.
- Tanaka, T. On the origin, affinity and scientific name of the Satsuma orange (2). Stud. Citrol. 1927, 1, 26–41. (In Japanese)
