As sessile organisms, flooding/submergence is one of the major abiotic stresses for higher plants, with deleterious effects on their growth and survival. Therefore, flooding/submergence is a large challenge for agriculture in lowland areas worldwide. Long-term flooding/submergence can cause severe hypoxia stress to crop plants and can result in substantial yield loss. Rice has evolved distinct adaptive strategies in response to low oxygen (O2) stress caused by flooding/submergence circumstances. Recently, direct seeding practice has been increasing in popularity due to its advantages of reducing cultivation cost and labor. However, establishment and growth of the seedlings from seed germination under the submergence condition are large obstacles for rice in direct seeding practice. The physiological and molecular regulatory mechanisms underlying tolerant and sensitive phenotypes in rice have been extensively investigated. Here, this review focuses on the progress of recent advances in the studies of the molecular mechanisms and metabolic adaptions underlying anaerobic germination (AG) and coleoptile elongation. Further, we highlight the prospect of introducing quantitative trait loci (QTL) for AG into rice mega varieties to ensure the compatibility of flooding/submergence tolerance traits and yield stability, thereby advancing the direct seeding practice and facilitating future breeding improvement.
- submergence
- direct seeding
- anaerobic germination
- low O2 stress
- regulatory mechanism
- metabolic adaptation
1. Introduction
Rice (Oryza sativa L.) is a staple food crop feeding more than half of the world’s population [1]. The traditional rice production system in Asian countries commonly involves transplanting seedlings from the nursery into a paddy field. This production pattern is labor-, water-, and energy-consuming and is becoming less profitable [2]. Therefore, in recent years, direct seeding has been receiving much attention worldwide, especially in Asian countries, and farmers have shifted to direct seeding of rice due to its low cost and labor-saving strategy [2][3][4]. There are three major methods for rice direct seeding: dry seeding (sowing dry seeds into dry soil), wet seeding (sowing pregerminated seeds on wet puddled soils), and water seeding (sowing seeds into standing water) [5]. However, poor crop stand establishment has become a key obstacle that prevents the subsequent growth, development, and yield in the direct seeding of rice [5]. In particular, water seeding severely affects the establishment of rice seedlings due to low water oxygen under the submergence condition.
For the early emerging seedlings, submergence reduces air diffusion, which limits O2 and carbon dioxide (CO2) availability in the submerged tissues, resulting in a hypoxic (<21% O2) environment, with an O2 concentration below that under normoxic conditions [6][7][8][9]. At low O2 supply (hypoxia) conditions, O2 concentration can affect the respiration in plants, and respiration can still active although at a strong reduced rate when O2 concentration is below the critical oxygen pressure for respiration (COPR), which is the lowest oxygen partial pressure to support maximum respiration rate [10][11]. Under O2 deficiency conditions, the accumulation of several phytotoxic substances such as reduced iron (Fe2+), manganese (Mn2+), hydrogen sulfide (H2S), oxygen radicals, and the products of fermentation, cause severe damage to plants. The ability of rice seeds to avoid the damages caused by these toxins is helpful to prevent further injury [12]. Although most modern rice varieties have low ability to germinate under water, rice has developed various adaptive mechanisms morphologically and physiologically in order to adapt to a wide range of hydrological environments. There are genetic variations exist among rice varieties, and these variations lay the base for the possibility of studying the molecular regulatory mechanism of AG-related traits and facilitate breeding rice varieties that are suitable for direct seeding systems . In direct seeding practice, varieties that are suitable for water seeding have characteristics of high anaerobic germination tolerance, early vigorous seedling growth, fast root growth, early tillering, and lodging resistance [13]. Therefore, the breeding of rice varieties that are capable of surviving and accommodating submergence conditions during germination and early growth stages improves the success of direct-seeded rice [14]. Thus, prior to breeding applications, it is of great importance to isolate related genes for AG tolerance from tolerant genotypes and to understand the genetic basis and the molecular regulatory mechanisms underlying anaerobic germination and submerged seedling growth.
Despite the sophisticated molecular mechanisms involved in the regulation of the anaerobic germination and seedling growth in rice, considerable progress on understanding the genetic, molecular and physiological basis of rice in response to submergence stress at the germination stage, has been achieved in the last few decades [15][16][17][18][19][20][21][22][23]. Here, we mainly focus on reviewing the molecular genetic mechanisms that regulate the metabolic adaptation of rice to anaerobic germination stress; we could not cover all the related advances due to space limitations. The advances reviewed here are crucial for understanding the fundamental mechanisms and for the breeding design in direct seeding practice. Last but not least, we proposed future research prospects and premised the issues raised that remain for further investigation.
2. Strategies are Adopted in Rice under Submerged Germination
Anaerobic germination (AG) tolerance is an important trait required for rice to successfully germinate in a direct seeding system. Rice has the ability to perceive low O2 stress and regulate germination and early seedling growth under hypoxic conditions [23][24][25]. There are two distinct adaptive responses or survival strategies for coping with submergence stress in plants: one is low O2 escape strategy (LOES), and the other is low O2 quiescence strategy (LOQS) [26][27][28][29]. In the case of the established rice plants, the two adaptive strategies for flooding tolerance are SUBMERGENCE 1A (SUB1A)-dependent quiescence strategy and SNORKEL 1 and SNORKEL 2 (SK1/2)-dependent escape strategy [30]. Overexpression of SUB1A stimulated the expression of alcohol dehydrogenase (ADH) genes for ethanolic fermentation and conferred submergence tolerance at the vegetative stage in rice by up-regulating the expression of slender rice-1 (SLR1) and SLR1 like-1(SLRL1), two negative regulators for gibberellin (GA) signaling, thereby inhibiting rice elongation, reducing the carbohydrate consumption, and exhibiting a strong submergence tolerance in rice plants. The rice varieties with SUB1A gene could survive up to two weeks of the complete submergence [31][32][33]. For SNORKEL 1 and SNORKEL 2 (SK1/2)-dependent escape strategy, SK1/2 promotes the internode elongation via the stimulation of GA biosynthesis in deep-water rice under flooding stress, and thereby enabling rice grows upward to the water surface for air exchange [34].
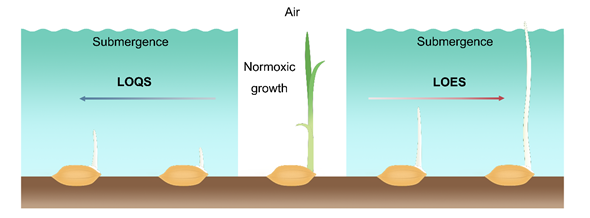
During germination and early seeding stages of rice, the LOES is characterized by promoting the elongation of the mesocotyl and coleoptile to allow plants to reach the water surface to increase air exchange between the aerial and submerged tissues. In rice, varieties that have adopted LOES in response to the submergence stress during germination are considered stress-tolerant. Conversely, LOQS is a strategy that constrains the elongation of the mesocotyl and coleoptile and preserves energy for prolonged submergence stress. The varieties that exhibit slow coleoptile growth under low O2 conditions are sensitive to submergence stress, thereby impairing the subsequent growth and development of the seedlings (Figure 1). Therefore, coleoptile elongation is a main index for selection of anerobic germination-tolerant rice varieties.
Figure 1. Low O2 escape strategy (LOES) and low O2 quiescence strategy (LOQS) strategy of rice germination under submergence conditions. Oxygen is one of the important factors that affects the germination and early seedling growth of rice. Under normoxic conditions, the surrounding oxygen ensures normal seed germination and early growth of the seedlings. Under the submergence condition, the formation of roots is inhibited in rice seedlings after germination. Sensitive cultivars adopt the quiescence strategy that inhibits germination and coleoptile elongation under low O2 stress, while tolerant varieties show phenotypes with faster coleoptile elongation capacity to reach the water surface for air exchange.
Previous studies have revealed that cell elongation contributes to the rapid increase in the length of the coleoptile. During the early stage of germination, cell division is activated to increase cell numbers for coleoptile growth, and the subsequent cell expansion contributes to elongation of the coleoptile under anerobic conditions [35][36]. Some specific expansins were reported to contribute to coleoptile elongation in rice under anaerobic conditions [37][38]; for example, EXPA2, EXPA4, EXPA1, EXPB11, and EXPB17 have been reported to be highly expressed in the coleoptiles under submergence stress [39]. To adapt to the submergence environment during the germination and early stages of growth, rice plants have evolved specific regulatory mechanisms to modulate metabolic shifts in response to low O2 stress [40][41], which will be described in more detail in the following sections.
3. Metabolic Adaption to Anaerobic Germination in Rice
In the submerged germination of rice, aerobic respiration is severely inhibited due to water O2 deficiency, leading to differential expression of the genes that contribute to activating the essential mechanisms and, in turn, to shifting the metabolic processes to anerobic respiration, which is related to energy production and utilization in the germinating seeds [42]. These alternative pathways to aerobic respiration are known as fermentation, including alcoholic fermentation, lactic acid fermentation, and the alanine pathway [43][44][45]. Meanwhile, tricarboxylic acid (TCA) cycle and oxidative phosphorylation processes are inhibited, and the production of ATP is shifted from electron transport chains (ETCs) to glycolysis and ethanol fermentation. Consequently, the net energy yield from anaerobic fermentation (2 mol ATP per mol of glucose) is lower than that from aerobic respiration (30–32 mol ATP per mol of glucose) (Figure 2). Although limited amounts of energy are produced from ethanol fermentation, it is extremely important for the germination and seedling growth under submergence condition. The energy is allocated to critical bioprocesses, especially protein synthesis in the cells of the coleoptile, contributing to submergence tolerance and success in the establishment of seedlings [46]. Likewise, the reoxidization of NADH to NAD+ in anerobic fermentation is also required for continuous glycolysis [47]. A recent report suggested that the AG-tolerant genotypes of rice adopt a strategy that strongly increases starch degradation in the endosperm into metabolizable sugars, thereby supplying substrates for subsequent glycolysis and alcohol fermentation to produce the energy required for enhancing the germination vitality and for the rapid outgrowth of the coleoptile .
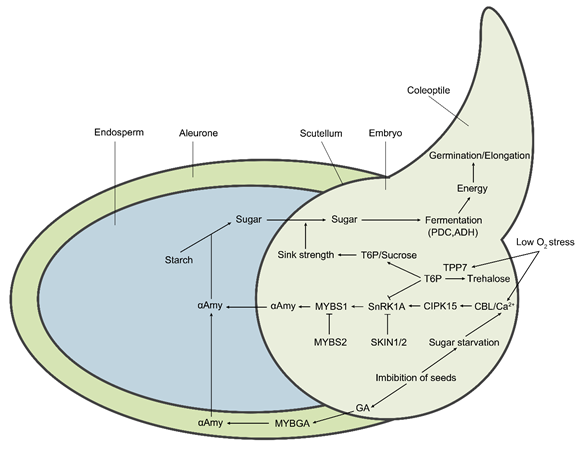
Figure 2. The molecular regulatory pathways and metabolic pathways of rice under hypoxic germination and early seedling growth. Sugar starvation caused by low O2 stress under the submergence condition is the key upstream signal affecting metabolic regulation pathways. The Ca2+ signal acts as a secondary messenger to mediate the downstream responses. The calcineurin B-like (CBL) proteins bind to Ca2+ and then interact with CIPK15, leading to the activation of its kinase activity. Subsequently, the activated CIPK15 physically interacts with SnRK1A, an upstream protein kinase of the transcription factor MYBS1, and activates its activity, consequently elevating the activity of αAmy for seed stored starch degradation. SKIN1/2 and MYBS2 negatively regulate SnRK1A and MYBS1, respectively, thereby repressing αAmy expression. Additionally, seed imbibition also results in the biosynthesis of gibberellin (GA) in the embryo. GA induces the expression of MYBGA in cereal aleurone cells, thereby upregulating the expression of the αAmy gene. Meanwhile, low O2-induced OsTPP7 desuppresses the SnRK1A activity that is inhibited by trehalose 6-phosphate (T6P). OsTPP7 increases the sink strength of the embryo axis–coleoptile by the perception of low sugar availability by enhancing the conversion of T6P to trehalose and leads to a decrease in the T6P/sucrose ratio, thus enhancing starch mobilization for energy production to promote coleoptile elongation. In addition, low O2 results in the shift of aerobic respiration to anaerobic fermentation, therefore inducing the expression of essential components, including PDC and ADH.
In the starch metabolic pathways, α-amylase (αAmy) (1,4-α-d-glucan maltohydrolase) is one of the most abundant hydrolases in rice and is involved in mobilization of the stored starch [48][49]; it catalyzes the hydrolysis of α-1,4-glucosidic bonds of starch, yielding α-glucose, and α-maltose [50]. In rice, a total of 10 distinct αAmy genes are classified into three subfamilies: subfamilies RAmy1 (A, B, and C), RAmy2A, and RAmy3 (A, B, C, D, E, and F). Most of the αAmy genes belong to the RAmy3 subfamily [51][52], in which only one gene (LOC_Os08g36910) is induced by sugar starvation and O2 deficiency [53]. Thus, RAmy3D is considered an important enzyme in the fermentative metabolism pathway for metabolic shift regulation and in the production of energy in response to submergence stress (Table 1). Regarding the alcoholic fermentation process, ADH, pyruvate decarboxylase (PDC), and aldehyde dehydrogenase (ALDH) are key enzymes that are involved in the reduction of pyruvate into alcohol. ADH catalyzes the rate-limiting step of ethanol metabolism and is considered to be essential for carbohydrate metabolism that is critical for the germination and growth of rice under low O2 stress [54][55]. It has been reported that a mutation in rice ADH1 leads to a reduction in the level of ADH protein, and thus compromises the tolerance to anaerobic stress of the adh-null mutant, suggesting that a functional ADH protein is required for enhancing tolerance to submergence during rice germination [56] (Figure 2). Moreover, pyruvate is a final product of glycolysis, which is converted to lactate by lactate dehydrogenase (LDH). The reduction in cytoplasmic pH inhibits the activity of LDH and activates the activity of PDC, leading to the reduction of intracellular lactate concentration. Therefore, the regulation of cytoplasmic pH, enables rice to avoid acidosis and improves the survival under low O2 conditions. Therefore, enhancement of the activity of major enzymes for either starch hydrolysis or ethanol synthesis is in favor of promoting the elongation of rice coleoptile under low O2 stress [[57].
Table 1. Subfamilies of the rice αAmy genes.[58][59][60]
|
Gene |
Subfamily |
Chromosome |
Locus Name |
CDS Coordinates (5’-3’) |
Regulatory Function |
||
|
RAmy1 A |
RAmy1 |
2 |
LOC_Os02g52700 |
32243146-32245056 |
High temperature in developing seeds [58] |
||
|
RAmy1 B |
2 |
LOC_Os02g52710 |
32250180-32248279 |
Chemical inhibition [59] |
|||
|
RAmy1 C |
1 |
LOC_Os01g25510 |
14459951-14461849 |
High temperature in developing seeds [58] |
|||
|
RAmy2 A |
RAmy2 |
6 |
LOC_Os06g49970 |
30262778-30266915 |
Unknown |
||
|
RAmy3 A |
RAmy3 |
9 |
LOC_Os09g28400 |
17288993-17291295 |
High temperature in developing seeds [58] |
||
|
RAmy3 B |
9 |
LOC_Os09g28420 |
17296166-17305076 |
Chemical inhibition [59] |
|||
|
RAmy3 C |
8 |
LOC_Os08g36900 |
23335165-23337151 |
Unknown |
|||
|
RAmy3 D |
8 |
LOC_Os08g36910 |
23340676-23343533 |
High temperature in developing seeds [58] Sugar starvation [53,60] Calcium signaling [21] |
|||
|
RAmy3 E |
4 |
LOC_Os04g33040 |
20006128-2000927 |
High temperature in developing seeds [58] Chemical inhibition [59] |
|||
|
RAmy3 F |
1 |
LOC_Os01g51754 |
29760719-29770037 |
Unknown |
|||
Upon imbibition of cereal grains, the rapid consumption of soluble carbohydrates during the germination and seedling growth stages causes sugar starvation and the activation of αAmy biosynthesis in the scutellum, which is mediated by the CIPK15-SnRK1A-MYBS1 signaling pathway [61]. Among major cereals, only rice produces a complete set of enzymes to degrade starch under AG . With a higher αAmy activity for the degradation of starch to produce energy in germinating seeds, rice promotes coleoptile elongation for surviving the anerobic stress .
4. The Molecular Regulatory Pathway of Anaerobic Germination in Rice
As mentioned above, rice submergence-tolerant varieties adopt LOES in response to low O2 stress. In submerged germination, O2 deficiency and sugar starvation are the key signals for rice responsiveness. O2 is one of the key components for the important biochemical reactions in rice cells. When rice germinates under submergence conditions, it senses hypoxic stress and induces the responsive regulatory pathways to adapt to the adverse environment. Calcium (Ca2+) signal, acting as a secondary messenger, is involved in multiple plant abiotic/biotic stress responsive signaling pathways [62]. The calcineurin B-like (CBL) proteins, a family of Ca2+-binding proteins and one of the major calcium ion sensors in plants, interact with CBL protein-interacting protein kinases (CIPKs) to form the CBL–CIPK complex, thereby decoding Ca2+ signaling in response to various abiotic stresses [63][64][65]. CIPKs is a group of plant-specific Ser/Thr protein kinases that contain an N-terminal kinase catalytic domain and a C-terminal regulatory NAF (Asn-Ala-Phe) domain. The NAF domain of CIPKs is sufficient for interaction with CBL protein and leads to the activation of the CIPKs [66].
Recent studies have revealed that the CBL/CIPK15-SnRK1A-MYBS1-mediated sugar-signaling cascade plays an important role in low-O2 tolerance during the seed germination stage of rice . CIPK15 is essential for regulating the expression of genes encoding αAmy and ADH (Figure 2). CBL4 has been reported to be induced by low-oxygen conditions and strongly interacts with the CIPK15 protein. The silence of CBL4 results in downregulation of the expression of αAmy3 (RAmy3 D). These findings suggest that the CBL4/CIPK15 complex plays a critical role in the sugar starvation signaling pathway and may contribute to the germination of rice under the hypoxia condition [67]. OsCBL10, another member of the rice CBL family, has been reported to be involved in flooding stress response during rice germination by comparing flooding-tolerant (Low88) with flooding-intolerant (Up221) cultivars. The natural variation in the OsCBL10 promoter is associated with the tolerance of anaerobic germination and seedling growth among rice subspecies; according to the sequence variation, its promoter can be characterized into two types, including a flooding-tolerant type (T-type) that only exists in japonica lowland cultivars and a flooding-intolerant type (I-type) that exists in japonica upland-, and indica upland/lowland-, cultivars. The flooding-intolerant cultivars that contain an I-type promoter can downregulate CIPK15 protein accumulation by inducing the expression of OsCBL10. In contrast, the cultivars containing a T-type promoter have downregulated OsCBL10 transcription, higher αAmy activity, and increased CIPK15 protein accumulation. Furthermore, however, the OsCBL10-overexpressing line is more sensitive to flooding stress than the wild type, suggesting that CBL10 negatively regulates the CIPK15-SnRK1A-MYBS1-mediated pathway.
Rice sucrose nonfermenting-1 (SNF1) homolog SNF1-related protein kinase-1 (SnRK1) is encoded by SnRK1A and SnRK1B, and SnRK1A is structurally and functionally analogous to SNF1 in yeast and AMP-activated protein kinase (AMPK) in mammals [68]. SnRK1A, SNF1, and AMPK are considered as metabolic sensors for monitoring cellular carbohydrate status and/or AMP/ATP levels, play an important role in the regulation of carbon and nitrogen metabolism, and are essential for the proper growth and development of higher eukaryotes [68][69][70]. In the downstream of CBL/CIPK15 complex, SnRK1A is involved in sugar starvation and O2 deficiency signaling pathways and acts as a conserved energy and stress sensor under submergence condition. Additionally, SnRK1A-interacting negative regulators (SKINs) physically interact with SnRK1A and negatively regulate its activity to modulate the nutrient starvation signaling and its downstream pathways, including inhibition of the expression of MYBS1 and αAmy 3, consequently resulting in compromising anaerobic germination and seedling growth under submergence stress in rice (Figure 2).
Myeloblastosis sugar response complex 1 (MYBS1) and MYBGA are two crucial transcription factors that can integrate diverse nutrient starvation and GA signaling pathways in downstream of SnRK1A in rice [71]. Under sugar-depleted conditions, MYBS1 specifically binds to the TATCCA cis-acting element (the TA-box) that is located on the sugar response complex (SRC) of the αAmy promoter, and consequently elevates the activity of αAmy for hydrolysis of starch in the endosperm [72][73]. SnRK1A mediates the interaction between MYBS1 and αAmy 3 SRC, and thus regulates the seed germination and seedling growth of rice (Figure 2). In addition, the phytohormone GA controls diverse aspects of plant growth and development, including seed germination and stem elongation [74]. Endogenous GA is synthesized in embryo and is then transported to the aleurone cells to promote the secretion of αAmy[75][76][77]. Furthermore, MYBGA is a GA-inducible MYB transcription factor that is expressed in cereal aleurone cells [78]. With GA induction, MYBGA upregulates the expression of αAmy by binding to the GA-responsive element (GARE) of the αAmy 3 promoter [79][80]. The formation of a stable bipartite MYB (MYBS1-MYBGA)-DNA (GARE-TA box) complex in the cytoplasm coordinates the expression of αAmy genes [71]. A recent study revealed that the transcription factor MYBS2 competes with MYBS1 for the same cis-element of the αAmy promoter and represses αAmy expression (Figure 2). Via promoting or inhibiting the expression level of αAmy, the competition between MYBS1 and MYBS2 can regulate and maintain sugar within an appropriate range during germination and early seedling growth under sugar starvation stress .
Moreover, trehalose-6-phosphate (T6P) metabolism is modulated in response to submergence stress. T6P phosphatase catalyzes the conversion of T6P to trehalose and changes the T6P:sucrose ratio [81]. The elevated level of T6P represses the activity of SnRK1A [82] (Figure 2). Therefore, T6P, is considered a sensor of sucrose effectiveness under low oxygen stress and is critical for allocating carbon from source to sink tissues and for maintaining sucrose homeostasis, which influences many metabolic and developmental processes in plants [83][84][.
In addition, phytohormones also play an important role in rice under submergence. Ethylene has a regulatory role in the upstream of both the SUB1A-dependent quiescence strategy and SK1/2-dependent escape strategy [85]. GA plays negative roles in SUB1A-dependent pathway mediating submergence tolerance and plays positive roles in the SK1/2-dependent pathway, triggering rapid internode elongation. GA also promote the elongation of rice coleoptile in the CIPK15-dependent pathways under submergence [86].
Thus, CIPK15 is induced by sugar starvation/low O2 signals caused by submergence and then activates the accumulation of SnRK1A protein. The accumulation of SnRK1A triggers a SnRK1A-dependent signaling cascade in response submergence stress. The downstream SnRK1A-MYBS1-αAmy 3 sugar signaling cascade promotes starch degradation and sugar mobilization, indicating that the CIPK15-SnRK1A-MYBS1-mediated sugar sensing pathway plays a key role in the submerged germination of rice seeds.
This entry is adapted from the peer-reviewed paper 10.3390/plants9101363
References
- Khush, G. Productivity Improvements in Rice. Nutr. Rev. 2003, 61 (Suppl. 6), S114–S116.
- Kumar, V.; Ladha, J.K. Direct Seeding of Rice. Recent Developments and Future Research Needs. Adv. Agron. 2011, 111, 297–413.
- Pandey, S.; Velasco, M.L. Economics of direct seeding in Asia: Patterns of adoption and research priorities. IRRI 1999, 24, 6–11.
- Miro, B.; Ismail, A.M. Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.). Front. Plant Sci. 2013, 4, 269.
- Farooq, M.; Siddique, K.H.M.; Rehman, H.; Aziz, T.; Lee, D.-J.; Wahid, A. Rice direct seeding: Experiences, challenges and opportunities. Soil Till. Res. 2011, 111, 87–98.
- Gibbs, J.; Greenway, H. Review: Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol. 2003, 30, 353–353.
- Sasidharan, R.; Bailey-Serres, J.; Ashikari, M.; Atwell, B.J.; Colmer, T.D.; Fagerstedt, K.; Fukao, T.; Geigenberger, P.; Hebelstrup, K.H.; Hill, R.D.; et al. Community recommendations on terminology and procedures used in flooding and low oxygen stress research. New Phytol. 2017, 214, 1403–1407.
- Sasidharan, R.; Hartman, S.; Liu, Z.; Martopawiro, S.; Sajeev, N.; van Veen, H.; Yeung, E.; Voesenek, L. Signal Dynamics and Interactions during Flooding Stress. Plant Physiol. 2018, 176, 1106–1117.
- Voesenek, L.A.; Sasidharan, R.; Visser, E.J.; Bailey-Serres, J. Flooding stress signaling through perturbations in oxygen, ethylene, nitric oxide and light. New Phytol. 2016, 209, 39–43.
- Wegner, L.H. Oxygen Transport in Waterlogged Plants. In Waterlogging Signalling and Tolerance in Plants; Springer: Berlin, Heidelberg, 2010; pp. 3–22.
- Armstrong, W.; Webb, T.; Darwent, M.; Beckett, P. Measuring and interpreting respiratory critical oxygen pressures in roots. Ann. Bot. 2008, 103, 281–293.
- Drew, M.; Lynch, J. Soil Anaerobiosis, Microorganisms, and Root Function. Annu. Rev. Phytopathol. 1980, 18, 37–66.
- Hafeez-ur-Rehman; Nawaz, A.; Awan, M.I.; Ijaz, M.; Hussain, M.; Ahmad, S.; Farooq, M. Direct Seeding in Rice: Problems and Prospects. In Agronomic Crops: Volume 1: Production Technologies; Hasanuzzaman, M., Ed.; Springer: Singapore, 2019; pp. 199–222.
- Pathak, D.S.; Tewari, A.; Sankhyan, S.; Dubey, D.; Mina, U.; Kumar, V.; Jain, N.; Bhatia, A. Direct-seeded rice: Potential, performance and problems-A review. Curr. Adv. Agric. Sci. 2011, 3, 77–88.
- Lee, K.-W.; Chen, P.-W.; Lu, C.-A.; Chen, S.; Ho, T.-H.; Yu, S.-M. Coordinated Responses to Oxygen and Sugar Deficiency Allow Rice Seedlings to Tolerate Flooding. Sci. Signal. 2009, 2, ra61.
- Ye, N.H.; Wang, F.Z.; Shi, L.; Chen, M.X.; Cao, Y.Y.; Zhu, F.Y.; Wu, Y.Z.; Xie, L.J.; Liu, T.Y.; Su, Z.Z.; et al. Natural variation in the promoter of rice calcineurin B-like protein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies. Plant J. 2018, 94, 612–625.
- Lu, C.A.; Lin, C.C.; Lee, K.W.; Chen, J.L.; Huang, L.F.; Ho, S.L.; Liu, H.J.; Hsing, Y.I.; Yu, S.M. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 2007, 19, 2484–2499.
- Kretzschmar, T.; Pelayo, M.A.; Trijatmiko, K.R.; Gabunada, L.F.; Alam, R.; Jimenez, R.; Mendioro, M.S.; Slamet-Loedin, I.H.; Sreenivasulu, N.; Bailey-Serres, J.; et al. A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 2015, 1, 15124.
- Guglielminetti, L.; Yamaguchi, J.; Perata, P.; Alpi, A. Amylolytic Activities in Cereal Seeds under Aerobic and Anaerobic Conditions. Plant Physiol. 1995, 109, 1069–1076.
- Matsumura, H.; Takano, T.; Yoshida, K.; Takeda, G. A Rice Mutant Lacking Alcohol Dehydrogenase. Jpn. J. Breed. 1995, 45, 365–367.
- Ho, V.T.; Tran, A.; Cardarelli, F.; Perata, P.; Pucciariello, C. A calcineurin B-like protein participates in low oxygen signalling in rice. Funct. Plant Biol. 2017, 44, 917–928.
- Lin, C.R.; Lee, K.W.; Chen, C.Y.; Hong, Y.F.; Chen, J.L.; Lu, C.A.; Chen, K.T.; Ho, T.H.; Yu, S.M. SnRK1A-interacting negative regulators modulate the nutrient starvation signaling sensor SnRK1 in source-sink communication in cereal seedlings under abiotic stress. Plant Cell 2014, 26, 808–827.
- Chen, Y.-S.; David Ho, T.-H.; Liu, L.; Lee, D.H.; Lee, C.-H.; Chen, Y.-R.; Lin, S.-Y.; Lu, C.-A.; Yu, S.-M. Sugar starvation-regulated MYBS2 and 14-3-3 protein interactions enhance plant growth, stress tolerance, and grain weight in rice. Proc. Natl. Acad. Sci. USA 2019, 116, 21925.
- Ismail, A.M.; Ella, E.S.; Vergara, G.V.; Mackill, D.J. Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa). Ann. Bot. 2009, 103, 197–209.
- Magneschi, L.; Perata, P. Rice germination and seedling growth in the absence of oxygen. Ann. Bot. 2009, 103, 181–196.
- Voesenek, L.A.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73.
- Bailey-Serres, J.; Voesenek, L.A. Flooding Stress: Acclimations and Genetic Diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339.
- Bailey-Serres, J.; Voesenek, L.A.C.J. Life in the balance: A signaling network controlling survival of flooding. Curr. Opin. Plant Biol. 2010, 13, 489–494.
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flooding tolerance: O2 sensing and survival strategies. Curr. Opin. Plant Biol. 2013, 16, 647–653.
- Bailey-Serres, J.; Lee, S.C.; Brinton, E. Waterproofing crops: Effective flooding survival strategies. Plant Physiol. 2012, 160, 1698–1709.
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708.
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034.
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819.
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030.
- Atwell, B.J.; Waters, I.; Greenway, H. The Effect of Oxygen and Turbulence on Elongation of Coleoptiles of Submergence-Tolerant and -Intolerant Rice Cultivars. J. Exp. Bot. 1982, 33, 1030–1044.
- Takahashi, H.; Saika, H.; Matsumura, H.; Nagamura, Y.; Tsutsumi, N.; Nishizawa, N.K.; Nakazono, M. Cell division and cell elongation in the coleoptile of rice alcohol dehydrogenase 1-deficient mutant are reduced under complete submergence. Ann. Bot. 2011, 108, 253–261.
- Huang, J.; Takano, T.; Akita, S. Expression of α-expansin genes in young seedlings of rice (Oryza sativa L.). Planta 2000, 211, 467–473.
- Choi, D.; Lee, Y.; Cho, H.T.; Kende, H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 2003, 15, 1386–1398.
- Lasanthi-Kudahettige, R.; Magneschi, L.; Loreti, E.; Gonzali, S.; Licausi, F.; Novi, G.; Beretta, O.; Vitulli, F.; Alpi, A.; Perata, P. Transcript Profiling of the Anoxic Rice Coleoptile. Plant Physiol. 2007, 144, 218–231.
- Lee, K.W.; Chen, P.W.; Yu, S.M. Metabolic adaptation to sugar/O2 deficiency for anaerobic germination and seedling growth in rice. Plant Cell Environ. 2014, 37, 2234–2244.
- Kuroha, T.; Ashikari, M. Molecular mechanisms and future improvement of submergence tolerance in rice. Mol. Breed. 2020, 40, 41.
- Hsu, S.K.; Tung, C.W. RNA-Seq Analysis of Diverse Rice Genotypes to Identify the Genes Controlling Coleoptile Growth during Submerged Germination. Front. Plant Sci. 2017, 8, 762.
- Greenway, H.; Gibbs, J. Mechanisms of anoxia tolerance in plants. II. Energy requirements for maintenance and energy distribution to essential processes. Funct. Plant Biol. 2003, 30, 999–1036.
- Sousa, C.A.F.D.; Sodek, L. The metabolic response of plants to oxygen deficiency. Braz. J. Plant Physiol. 2002, 14, 83–94.
- Tadege, M.; Dupuis, I.; Kuhlemeier, C. Ethanolic fermentation: New functions for an old pathway. Trends Plant Sci. 1999, 4, 320–325.
- Edwards, J.M.; Roberts, T.H.; Atwell, B.J. Quantifying ATP turnover in anoxic coleoptiles of rice (Oryza sativa) demonstrates preferential allocation of energy to protein synthesis. J. Exp. Bot. 2012, 63, 4389–4402.
- Miro, B.; Longkumer, T.; Entila, F.D.; Kohli, A.; Ismail, A.M. Rice Seed Germination Underwater: Morpho-Physiological Responses and the Bases of Differential Expression of Alcoholic Fermentation Enzymes. Front. Plant Sci. 2017, 8, 1857.
- Damaris, R.N.; Lin, Z.; Yang, P.; He, D. The Rice Alpha-Amylase, Conserved Regulator of Seed Maturation and Germination. Int. J. Mol. Sci 2019, 20, 450.
- Senapati, S.; Kuanar, S.; Sarkar, R. Anaerobic Germination Potential in Rice (Oryza sativa L.): Role of Amylases, Alcohol deydrogenase and Ethylene. J. Stress Physiol. Biochem. 2019, 15, 39–52.
- Pujadas, G.; Palau, J. Evolution of alpha-amylases: Architectural features and key residues in the stabilization of the (beta/alpha)(8) scaffold. Mol. Biol. Evol. 2001, 18, 38–54.
- Huang, N.; Sutliff, T.D.; Litts, J.C.; Rodriguez, R.L. Classification and characterization of the rice alpha-amylase multigene family. Plant Mol. Biol. 1990, 14, 655–668.
- Huang, N.; Stebbins, G.; Rodriguez, R. Classification and evolution of α-amylase genes in plants. Proc. Natl. Acad. Sci. USA 1992, 89, 7526–7530.
- Hwang, Y.S.; Thomas, B.R.; Rodriguez, R.L. Differential expression of rice α-amylase genes during seedling development under anoxia. Plant Mol. Biol. 1999, 40, 911–920.
- Takahashi, H.; Greenway, H.; Matsumura, H.; Tsutsumi, N.; Nakazono, M. Rice alcohol dehydrogenase 1 promotes survival and has a major impact on carbohydrate metabolism in the embryo and endosperm when seeds are germinated in partially oxygenated water. Ann. Bot. 2014, 113, 851–859.
- Vijayan, J.; Senapati, S.; Ray, S.; Chakraborty, K.; Molla, K.; Basak, N.; Pradhan, B.; Yeasmin, L.; Chattopadhyay, K.; Sarkar, R. Transcriptomic and physiological studies identify cues for germination stage oxygen deficiency tolerance in rice. Environ. Exp. Bot. 2017, 147, 234–248.
- Saika, H.; Matsumura, H.; Takano, T.; Tsutsumi, N.; Nakazono, M. A Point Mutation of Adh1 Gene is Involved in the Repression of Coleoptile Elongation under Submergence in Rice. Breed. Sci. 2006, 56, 69–74.
- Kato-Noguchi, H.; Morokuma, M. Ethanolic fermentation and anoxia tolerance in four rice cultivars. J. Plant Physiol. 2007, 164, 168–173.
- Hakata, M.; Kuroda, M.; Miyashita, T.; Yamaguchi, T.; Kojima, M.; Sakakibara, H.; Mitsui, T.; Yamakawa, H. Suppression of α‐amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 2012, 10, 1110–1117.
- Hu, Q.; Fu, Y.; Guan, Y.; Lin, C.; Cao, D.; Hu, W.; Sheteiwy, M.; Hu, J. Inhibitory effect of chemical combinations on seed germination and pre-harvest sprouting in hybrid rice. Plant Growth Regul. 2016, 80, 281–289.
- Hwang, Y.S.; Karrer, E.E.; Thomas, B.R.; Chen, L.; Rodriguez, R.L. Three cis-elements required for rice α-amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 1998, 36, 331–341.
- Yu, S.M.; Lee, Y.C.; Fang, S.C.; Chan, M.T.; Hwa, S.F.; Liu, L.F. Sugars act as signal molecules and osmotica to regulate the expression of alpha-amylase genes and metabolic activities in germinating cereal grains. Plant Mol. Biol. 1996, 30, 1277–1289.
- Knight, H.; Knight, M. Abiotic stress signaling pathways: Specificity and cross-talk. Trends Plant Sci. 2001, 6, 1360–1385.
- Batistic, O.; Kudla, J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924.
- Das, R.; Pandey, G.K. Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr. Genom. 2010, 11, 2–13.
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42.
- Albrecht, V.; Ritz, O.; Linder, S.; Harter, K.; Kudla, J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001, 20, 1051–1063.
- Kurusu, T.; Hamada, J.; Nokajima, H.; Kitagawa, Y.; Kiyoduka, M.; Takahashi, A.; Hanamata, S.; Ohno, R.; Hayashi, T.; Okada, K.; et al. Regulation of Microbe-Associated Molecular Pattern-Induced Hypersensitive Cell Death, Phytoalexin Production, and Defense Gene Expression by Calcineurin B-Like Protein-Interacting Protein Kinases, OsCIPK14/15, in Rice Cultured Cells. Plant Physiol. 2010, 153, 678–692.
- Halford, N.G.; Hey, S.; Jhurreea, D.; Laurie, S.; McKibbin, R.S.; Paul, M.; Zhang, Y. Metabolic signalling and carbon partitioning: Role of Snf1-related (SnRK1) protein kinase. J. Exp. Bot. 2003, 54, 467–475.
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709.
- Ghillebert, R.; Swinnen, E.; Wen, J.; Vandesteene, L.; Ramon, M.; Norga, K.; Rolland, F.; Winderickx, J. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: Structure, function and regulation. FEBS J. 2011, 278, 3978–3990.
- Hong, Y.F.; Ho, T.H.; Wu, C.F.; Ho, S.L.; Yeh, R.H.; Lu, C.A.; Chen, P.W.; Yu, L.C.; Chao, A.; Yu, S.M. Convergent starvation signals and hormone crosstalk in regulating nutrient mobilization upon germination in cereals. Plant Cell 2012, 24, 2857–2873.
- Lu, C.A.; Ho, T.H.; Ho, S.L.; Yu, S.M. Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 2002, 14, 1963–1980.
- Lu, C.A.; Lim, E.K.; Yu, S.M. Sugar response sequence in the promoter of a rice α-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 1998, 273, 10120–10131.
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251.
- Chen, P.W.; Chiang, C.M.; Tseng, T.H.; Yu, S.M. Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes. Plant Cell 2006, 18, 2326–2340.
- Chen, P.W.; Lu, C.A.; Yu, T.S.; Tseng, T.H.; Wang, C.S.; Yu, S.M. Rice alpha-amylase transcriptional enhancers direct multiple mode regulation of promoters in transgenic rice. J. Biol. Chem. 2002, 277, 13641–13649.
- Kaneko, M.; Itoh, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M. The alpha-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 2002, 128, 1264–1270.
- Tsuji, H.; Aya, K.; Ueguchi-Tanaka, M.; Shimada, Y.; Nakazono, M.; Watanabe, R.; Nishizawa, N.K.; Gomi, K.; Shimada, A.; Kitano, H.; et al. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 2006, 47, 427–444.
- Gubler, F.; Kalla, R.; Roberts, J.K.; Jacobsen, J.V. Gibberellin-Regulated Expression of a myb Gene in Barley Aleurone Cells: Evidence for Myb Transactivation of a High-pl a-Amylase Gene Promoter. Plant Cell 1995, 7, 1879.
- Gubler, F.; Raventos, D.; Keys, M.; Watts, R.; Mundy, J.; Jacobsen, J. Target genes and regulatory domains of the GAMYB transcription activator in cereal aleurone. Plant J. 1999, 17, 1–9.
- Lunn, J.; Delorge, I.; Figueroa, C.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567.
- Zhang, Y.; Primavesi, L.F.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.A.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 2009, 149, 1860–1871.
- Figueroa, C.M.; Lunn, J.E. A Tale of Two Sugars: Trehalose 6-Phosphate and Sucrose. Plant Physiol. 2016, 172, 7–27.
- Yadav, U.P.; Ivakov, A.; Feil, R.; Duan, G.Y.; Walther, D.; Giavalisco, P.; Piques, M.; Carillo, P.; Hubberten, H.M.; Stitt, M.; et al. The sucrose-trehalose 6-phosphate (Tre6P) nexus: Specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 2014, 65, 1051–1068.
- Fukao, T.; Xiong, L. Genetic mechanisms conferring adaptation to submergence and drought in rice: Simple or complex? Curr. Opin. Plant Biol. 2013, 16, 196–204.
- Nagai, K.; Hattori, Y.; Ashikari, M. Stunt or elongate? Two opposite strategies by which rice adapts to floods. J. Plant Res. 2010, 123, 303–309.