Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Lung cancers are life-threatening malignancies that cause great healthcare burdens in Taiwan and worldwide. The 5-year survival rate for Taiwanese patients with lung cancer is approximately 29%, an unsatisfactorily low number that remains to be improved.
- Taiwan
- lung cancer
- precision medicine
- proteogenomics
- targeted therapy
- NRF2
1. Characteristics of Lung Cancer Patients in Taiwan
The lung is a vital organ responsible for the exchange of oxygen and carbon dioxides, and sustaining metabolism in humans. Lung cancer is the major cause of death for males and females with malignant tumors in Taiwan [1]. Only 15~20% of patients are diagnosed at early stages. Lung cancer comprises small cell lung cancer and non-small cell lung cancer (NSCLC), the predominant histology type, which is further categorized into adenocarcinoma, squamous cell carcinoma, large cell neuroendocrine carcinoma and others. The 5-year survival rate of Taiwanese patients with lung cancer has been reported to be approximately 28.6% in 2019. Of the ten most common malignancies in Taiwan, lung cancer has the lowest 5-year survival rate. The number of patients who died as a result of lung cancer increased from 5749 to 9701 between 1998 and 2019, suggesting that lung cancer is still a severe threat to the health and well-being of Taiwanese people [2]. The incidence of lung cancer in Taiwan has not decreased with time and the high mortality rate emphasizes the unmet need for optimizing treatment by leveraging a more personalized approach, including finding precise cancer subtypes, seeking driver oncogenes, overcoming drug resistance, and developing the optimal sequences of treatments [1].
2. Molecular Epidemiology and Pathophysiology of Lung Cancer in Taiwan
The distribution of patients with distinct molecular characteristics in a population, i.e., molecular epidemiology, is crucial for guiding the treatment policy to provide personalized treatments. NSCLC patients in Taiwan comprise higher percentages of nonsmokers than smokers, and females than males (Table 1). This is quite different from the demographics in western countries, such as the United States where there are more male patients and smokers (Table 1). Thus, molecular epidemiology of Taiwan warrants investigation. Recently, a comprehensive and deep proteogenomic resource of a cohort of treatment-naïve early-stage lung cancer patients in Taiwan was released, comprising high-quality molecular assessments of tumor and normal adjacent tissues (NAT) of surgical specimens, including gene expressions quantified by RNAseq and protein abundances quantified by mass spectrometry [3]. In this cohort, the majority of patients have adenocarcinoma. This resource facilitated the elucidation of lung cancer biology and molecular epidemiology in Taiwan.
Somatic mutation analysis of this cohort echoed a well-known fact that patients with the epidermal growth factor receptor (EGFR) mutations in the tumor account for 85% of the total patient population which comprises predominantly nonsmokers (83%) [3]. RBM10 and EGFR L858 mutations are more common in females, whereas KRAS and APC mutations are frequently found in male patients. Additionally, ATM and KRAS are often found to have mutated in smokers.
Gross functional evaluations of increasing and decreasing proteins in this deep proteogenomic resource was performed with respect to the tumor stages. The increasing proteins are associated with DNA replication, glycolysis and stress response. The decreasing proteins pertain to cell-to-cell communication, signaling, integrin, G protein coupled receptors, ion channels and adaptive immunity. Based on the protein profiles of the tumor microenvironment, patients were clustered into three subgroups. The first subgroup accounts for 61% of the cohort. This subgroup is associated with more advanced cancer stages and visceral pleural invasion, as well as higher mutation burdens. Subgroup 2 accounts for 12% of the cohort and is associated with EGFR L858R mutations. Subgroup 3 accounts for 27% of the cohort and is associated with PI3K/AKT pathways and cell cycles, identified by protein phosphorylation analysis. Moderate RNA-to-protein correlations are found in this research, resulting in a slight disparity in the patient subgroups detected using RNA and protein alone [3].
An independent analysis of this proteogenomic resource further elucidated that the nuclear factor erythroid 2-related factor 2 (NRF2)/nuclear factor erythroid-derived 2-like 2 (NFE2L2) antioxidant mechanisms underlie the oncogenesis and tumor progression of lung adenocarcinoma in Taiwan, based on the higher expressions of NRF2 antioxidant genes in the tumor than in adjacent normal tissues [5]. Data suggested that the NRF2 antioxidant mechanism was the most prominent mechanism, among a total of 189 oncogenic mechanisms evaluated. This was a gross analysis performed regardless of the three subgroups mentioned above, with the goal of discovering the shared driving mechanism. To cope with the carcinogens and/or oxidative stress caused by the fast division of cells, NRF2 antioxidant mechanisms are induced. NRF2 is a transcription factor that can regulate the expression of other genes via binding with the antioxidant-response elements (ARE) of the genome and activating a series of downstream effects (Figure 1). These activities altogether mediate the metabolic reprogramming and increased antioxidant capability of the cancer cell.
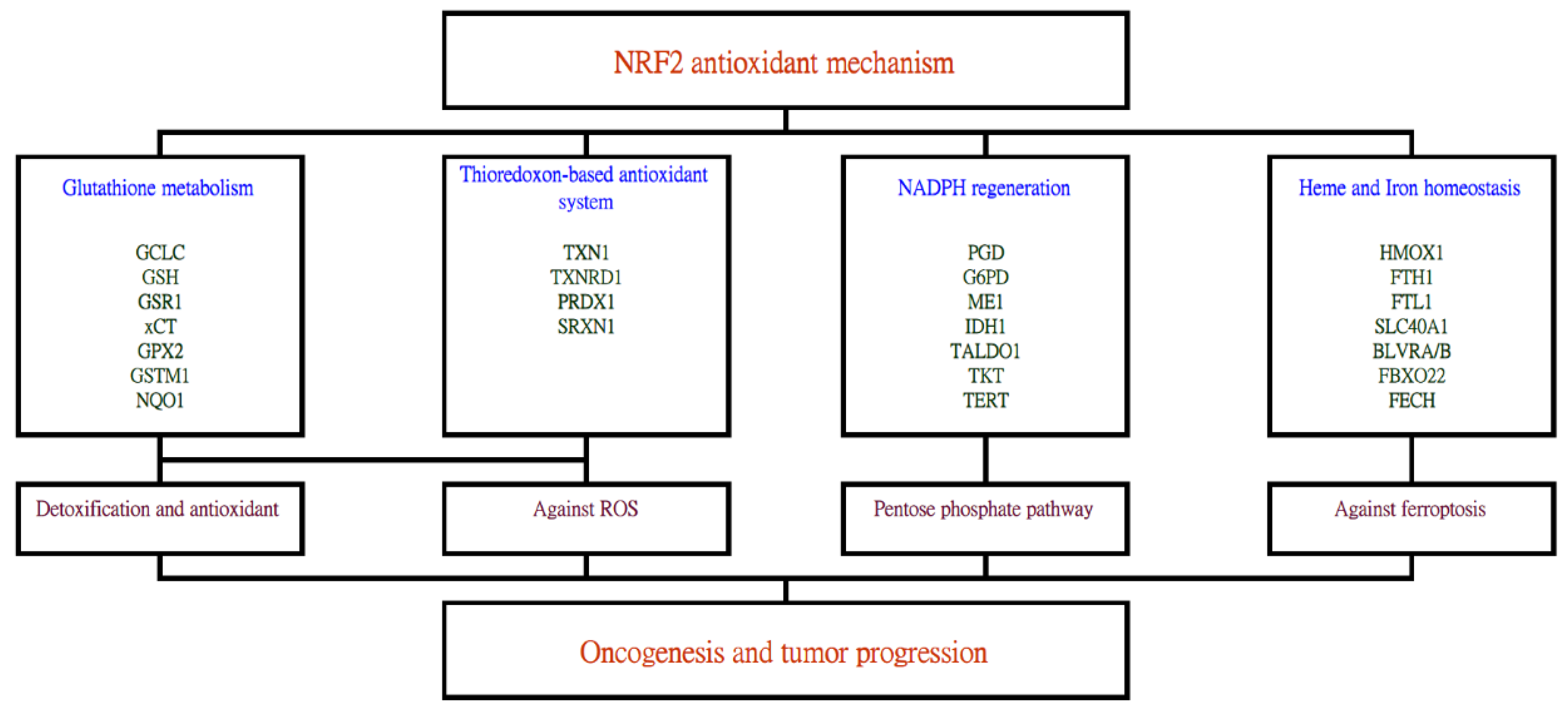
Figure 1. The stress-induced NRF2 antioxidant system can activate a series of downstream genes which ultimately leads toward lung cancer carcinogenesis and progression. Genes activated by the NRF2 transcription factor include those related to glutathione metabolism, thioredoxin-based antioxidant system, NADPH regeneration and heme and iron homeostasis. Subsequent effects, such as detoxification, reactive oxygen species removal, pentose phosphate pathway and inhibition of ferroptosis then follow.
Apart from the aforementioned oncogenic mechanisms derived from this proteogenomic resource, new insights can still be obtained using more advanced multi-omics analysis methods, such as similarity network fusing [6]. Furthermore, several important mechanisms are worth noting. Tissue fibrosis and inflammatory mechanisms have been found to be an important molecular signature associated with cancer stage and the poor prognosis of lung cancer [7]. This is a mechanism akin to wound healing. Patients with lung cancer often have metastasis to the brain. Thus, the cross-referencing of brain and lung cancer gene-expression signatures is warranted [8]. Additionally, eukaryotic initiation factors represent crucial members which can activate oncogenic pathways [9].
Like most cancers, lung adenocarcinoma is a heterogeneous collection of diseases. Partitioning a heterogeneous collection of diseases into relatively homogeneous subgroups allows for more precise estimation of patient outcomes and more precise subgroup-specific treatments [10]. The established oncogenic mechanisms, including EGFR mutation, anaplastic lymphoma kinase (ALK) fusion, ROS1 fusion and RET fusion, only account for certain proportions of the entire patient population [4]. Hence, the search for new subtypes that better characterize the patient population is warranted. In research conducted in Japan, a country that is geographically close to Taiwan, two subtypes of lung adenocarcinoma were found using hierarchical clustering methods analyzing the miRNA profiles in the tumor tissues. The two subtypes are driven by the dysregulation of miRNA miR-30d and miR-195, respectively. The first subtype represents less differentiated tissue and implies poorer survival. The second subtype represents well-differentiated tissue and implies better survival [11]. In another study utilizing The Cancer Genome Atlas (TCGA) data, three subtypes (high, medium and low immunity) were identified [12]. The high-immunity group has a better response to immunotherapy and chemotherapy.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23137037
References
- Luo, Y.H.; Chiu, C.H.; Scott Kuo, C.H.; Chou, T.Y.; Yeh, Y.C.; Hsu, H.S.; Yen, S.H.; Wu, Y.H.; Yang, J.C.; Liao, B.C.; et al. Lung Cancer in Republic of China. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2021, 16, 519–527.
- Health Promotion Administration; Ministry of Health and Welfare. Taiwan. 2019 Cancer Registry Annual Report. Available online: https://www.hpa.gov.tw/Pages/ashx/File.ashx?FilePath=~/File/Attach/14913/File_17794.pdf (accessed on 4 February 2022).
- Chen, Y.-J.; Roumeliotis, T.I.; Chang, Y.-H.; Chen, C.-T.; Han, C.-L.; Lin, M.-H.; Chen, H.-W.; Chang, G.-C.; Chang, Y.-L.; Wu, C.-T.; et al. Proteogenomics of non-smoking lung cancer in east asia delineates molecular signatures of pathogenesis and progression. Cell 2020, 182, 226–244.e217.
- Gillette, M.A.; Satpathy, S.; Cao, S.; Dhanasekaran, S.M.; Vasaikar, S.V.; Krug, K.; Petralia, F.; Li, Y.; Liang, W.W.; Reva, B.; et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 2020, 182, 200–225.e235.
- Liang, K.-H.; Wang, M.-L. Deep proteogenomic investigations elucidate the NRF2 antioxidant mechanism as a major driving mechanism of lung adenocarcinoma in Asia. J. Chin. Med. Assoc. 2021, 84, 766–771.
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337.
- Chang, H.Y.; Sneddon, J.B.; Alizadeh, A.A.; Sood, R.; West, R.B.; Montgomery, K.; Chi, J.-T.; van de Rijn, M.; Botstein, D.; Brown, P.O. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol. 2004, 2, E7.
- Cahoy, J.D.; Emery, B.; Kaushal, A.; Foo, L.C.; Zamanian, J.L.; Christopherson, K.S.; Xing, Y.; Lubischer, J.L.; Krieg, P.A.; Krupenko, S.A.; et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008, 28, 264–278.
- Ramírez-Valle, F.; Braunstein, S.; Zavadil, J.; Formenti, S.C.; Schneider, R.J. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 2008, 181, 293–307.
- Bild, A.H.; Yao, G.; Chang, J.T.; Wang, Q.; Potti, A.; Chasse, D.; Joshi, M.-B.; Harpole, D.; Lancaster, J.M.; Berchuck, A.; et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2005, 439, 353–357.
- Arima, C.; Kajino, T.; Tamada, Y.; Imoto, S.; Shimada, Y.; Nakatochi, M.; Suzuki, M.; Isomura, H.; Yatabe, Y.; Yamaguchi, T.; et al. Lung adenocarcinoma subtypes definable by lung development-related miRNA expression profiles in association with clinicopathologic features. Carcinogenesis 2014, 35, 2224–2231.
- Xu, F.; Chen, J.-X.; Yang, X.-B.; Hong, X.-B.; Li, Z.-X.; Lin, L.; Chen, Y.-S. Analysis of Lung Adenocarcinoma Subtypes Based on Immune Signatures Identifies Clinical Implications for Cancer Therapy. Mol. Ther. Oncolytics 2020, 17, 241–249.
This entry is offline, you can click here to edit this entry!
