Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Microbiology
Treatment of infections caused by bacteria has become more complex due to the increasing number of bacterial strains that are resistant to conventional antimicrobial therapy. A highly promising alternative appears to be bacteriophage (phage) therapy, in which natural predators of bacteria, bacteriophages, play a role. Although these viruses were first discovered in 1917, the development of phage therapy was impacted by the discovery of antibiotics, which spread more quickly and effectively in medical practice.
- bacteriophages
- drug-resistant bacteria
- phage therapy
1. Introduction
In recent decades, an increasing number of antibiotic-resistant bacterial strains have been reported. Therefore, conventional antibiotic therapy is often ineffective [1,2,3], leading to the search for new treatment options for bacterial infectious diseases. Several possibilities exist to overcome bacterial resistance to antibiotics; for example, the development of new effective antibiotics [4]; the testing of the antimicrobial effect of natural substances, such as herbal products [5,6], honey [7] or wine products [8]; the use of photodynamic inactivation [9] or the quorum-quenching approach [10]; or therapy that modulates the patient’s own immunity using immunomodulators of microbial origin or therapeutic autovaccines prepared from the patient’s own strains [11]. One of the most promising alternatives to antibiotic treatment is bacteriophage therapy, which uses natural enemies of bacteria, bacteriophages [12]. Bacteriophages, or phages, are viruses that control the growth and spread of their bacterial hosts. They are the most widespread entity in the biosphere and are found wherever bacteria live, for example, in salt waters, cold waters, hot springs, waste waters, or sewage [13,14,15]. They are also the most abundant component of the human microbiome, playing an important role in intestinal microbial composition and horizontal gene transfer [16,17,18].
2. Phage Biology
2.1. Phage Life Cycle
The first step during phage infection is adsorption of phages to the surface structures of bacteria by tail fibers or bacteriophage spikes [14]. Phages are capable of infecting only those bacteria that have the corresponding receptor, which determines the host spectrum of the phages. This spectrum is also limited by the defense mechanisms of bacteria against phages [18]. This interaction results in a different host specificity—some phages are strain-specific, whereas others are able to infect a wider range of bacterial species or even genera [19].
After successful adsorption, a pore is formed in the membrane of the infected bacterium and the phage genome is subsequently injected into its cytoplasm [13]. Phage replication may be performed in two basic ways [19,20]. In the lytic cycle, phages exploit the proteosynthetic apparatus of bacteria to synthesize their proteins, replicate phage nucleic acid (NA), assemble phage particles, and release themselves from the cell while disrupting the host bacterial cell [14]. During the release, phages use holins to perforate the cytoplasmic membrane and endolysins to destroy the cell wall membrane, thus lysing the bacterial cell [21].
In the lysogenic cycle, the phage genome is integrated into the bacterial chromosome in the form of a prophage. Such an integrated phage can persist in the genetic material of the bacterium for a long time, and replicate its nucleic acid together with the replication of the bacterial chromosome. Prophages may persist in this state until they switch to the lytic cycle [20]. Lysogeny may provide benefits to the bacterial cell because phages are able to construct gene-encoding products that are involved in antibiotic resistance or virulence factors that enhance the infectious process (e.g., genes for diphtheria toxin [22] or Panton–Valentin leukocidin [23]). Therefore, in the context of phage therapy, a lytic life cycle is desirable, in addition to the absence of genetic determinants of antimicrobial resistance and bacterial virulence in the genome of therapeutic phages [15]. Differences between the lytic and lysogenic life cycle of bacteriophages are shown in Figure 1.
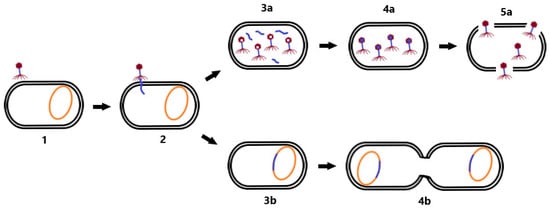
Figure 1. Life cycle of bacteriophages. First, the phage adsorbs to the bacterial cell receptor (1). Second, it forms a pore in the membrane and phage NA is injected into the host cell (2). As the virus continues the lytic cycle, viral proteins are produced, phage NA (3a) replicates, and virion assembly (4a) and cell lysis (5a) occur. During the lysogenic cycle, the phage integrates its genome into the host chromosome (3b) and replicates with it (4b). Changing conditions may induce a transition to the lytic cycle.
2.2. Phages versus Bacteria
During evolution, bacterial strains developed mechanisms to protect themselves from phage infection. One of these mechanisms is superinfection exclusion. In this case, the lysogenized bacterium that carries a prophage in its genome cannot be infected by another phage. Other mechanisms include preventing phage adsorption to the bacterial cell surface due to alteration of the receptor [18] or the production of protective surface polysaccharides that mask the receptor [24]. In addition, bacterial cells can reduce the number of potentially infectious bacteriophage particles by producing membrane vesicles [25]. If phage adsorption is successful, the host cell can subsequently prevent injection of phage NA by modification of the inner membrane proteins [26]. The infected bacterium can also identify the injected phage NA and degrade it via a restriction–modification system [27] or a Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated proteins system (CRISPR-Cas) [28]. Another possibility is preventing the replication of phage NA by secondary metabolites produced by the bacterial cell [29], or preventing the assembly of virions by inhibiting phage terminase or by scattering phage coat proteins [30]. If these bacterial cell defense mechanisms are not successful, the host cell may sacrifice itself for the benefit of others, resulting in cell death, and thus limiting the production of phage particles that may infect other bacterial cells in the environment (Figure 2) [31].
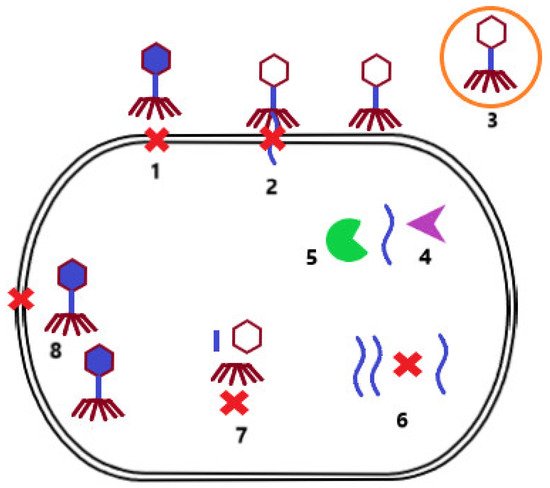
Figure 2. Mechanisms of bacterial resistance to phage infection. Prevention of phage adsorption to the surface of bacterial cells due to alteration of the surface receptor or production of protective surface polysaccharides (1). Prevention of phage NA injection by modification of inner membrane proteins (2). Reducing the number of free infectious bacteriophage particles by producing membrane vesicles (3). Degradation of phage NA by the restriction modification system (4) or the CRISP-Cas system (5). Prevention of phage NA replication by secondary bacterial metabolites (6). Inhibition of the assembly of phage particles by blocking terminase or scattering envelope proteins (7). Limitation of phage particle production by induction of cell death (8).
However, bacterial strains rarely have more than one defensive mechanism against phage infection [32]. Alternatively, phages may possess mechanisms that help overcome the defensive machinery of the host cell. These include, for example, the ability to produce glycosidases to degrade host capsules, and thus unmask receptors to initiate phage infection [33]. Another mechanism is the ability of phages to produce hypervariable receptor-binding proteins that allow binding to modified host receptors [18,34]. Phages can also overcome the CRISPR-Cas system by mutating or deleting target sites for this system or expressing proteins that interfere with CRISPR-Cas [35]. An important mechanism is the ability of phages to modify their NA to avoid the host restriction–modification system [36], or even the ability of phages to encode their own CRISPR-Cas system that interferes with host defensive mechanisms [37]. All of these mechanisms determine the specificity of phages against bacterial hosts [12].
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms10071365
This entry is offline, you can click here to edit this entry!
