Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The indolent nature of some cancers makes early detection challenging, as such significant effort is placed on identifying circulating cancer biomarkers using minimally invasive, highly sensitive diagnostic assays. Biological fluids contain small extracellular vesicles including exosomes, which have many tissue origins. Cancer cells increase production and release of exosomes in the circulation to deliver biologically active compounds that can reprogram recipient cells, which potentially represent a valuable source of biomarkers.
- Extracellular Vesicles
- Exosomes
- Circulating Biomarkers
- Next Generation Sequencing
- Mass Spectrometry
- Lipidomics
- Proteomics
- Transcriptomics
- Cancer
- Liquid Biopsy
1. Extracellular Vesicles (EVs): A Journey from Discovery to Clinical Utility
The first description of EVs can be traced back to 1946, to the studies of Erwin Chargaff and Randolph West on blood coagulation [1][2]. They were the first to describe a platelet-free coagulation component from plasma which could be sedimented into a pellet by high-speed centrifugation (31,000× g) and could inhibit blood clotting [3].
The definition of exosomes was introduced in complementary seminal papers from Philip Stahl and Rose Johnstone’s laboratories in 1983 [4][5]. Stahl published stunning electron microscopic images which demonstrated the mechanical process of exosome release from the lumen of MVBs upon fusion with the plasma membrane, which helped define this novel intracellular sorting pathway then referred to as the exosome secretion pathway [4]. Functionally, Rose Johnstone and co-workers suggested that exosomes served as a cellular waste disposal mechanism, wherein they showed that under different cellular stressors the presence of the transferrin receptor on exosomes was altered at different times, thus suggesting that exosomes provided a major route for the shedding of obsolete membrane proteins [6]. However, many contradicting papers were published demonstrating the lateral cell–cell diffusion/transfer of proteins and lipids within the membranes of exosomes (i.e., membrane fluidity) [7], along with the identification of functional enzymes [8] and other active components (i.e., the tetraspanins, Rab4, and ARF [9]), suggestive of their active biological function. Additionally, several papers demonstrated that the production of exosomes and their composition could be altered at different stages of disease, being increased during transient brain ischemia, myocardial infarctions, angina, and Crohn’s disease, wherein exosomes were shown to act as functional activators [10][11][12].
Functional studies, many of which were conducted in the early 2000s, have highlighted the importance of the exosome cargoes and their functions as unique biological devices. Exosome proteomics, lipidomics, and transcriptomics analyses have led to the discovery of unique exosome cell-specific cargoes [13][14][15][16], confirming that they transport active proteins and also, importantly, carry unique and intact RNA molecules (i.e., mRNAs, miRNAs, and lncRNAs) that can post-transcriptionally modify the fate and behavior of recipient cells [17][18][19]. The study by Skog et al. demonstrated that the RNA found in glioblastoma-derived exosomes contains a “snapshot” of the cellular transcriptome of their cells of origin at a specific point in time [20]. With an ever-growing number of studies demonstrating the powerful and oncogenic effects of tumor exosomes, important breakthroughs such as the mechanistic description of metastatic dissemination have highlighted the unique intercellular communication properties of exosomes [21][22][23][24]. Jang et al. demonstrated in their study that the enhanced release of breast tumor-derived exosomes is critical for the education of pre-metastatic niche cells, which accommodate the colonization of circulating cancer cells from primary tumors [25][26]. This has in part been explained by the “seed and soil” hypothesis proposed by Stephen Paget [27], wherein exosomes carry oncogenic signals (i.e., mRNA, miRNA, and protein) specifically packaged by primary tumors’ (i.e., the “seed”) to prime target sites (i.e., the “soil”) prior to the dissemination of tumor cells, a process by which secondary tumors can prosper in a suitable microenvironment [28][29] (Figure 1).
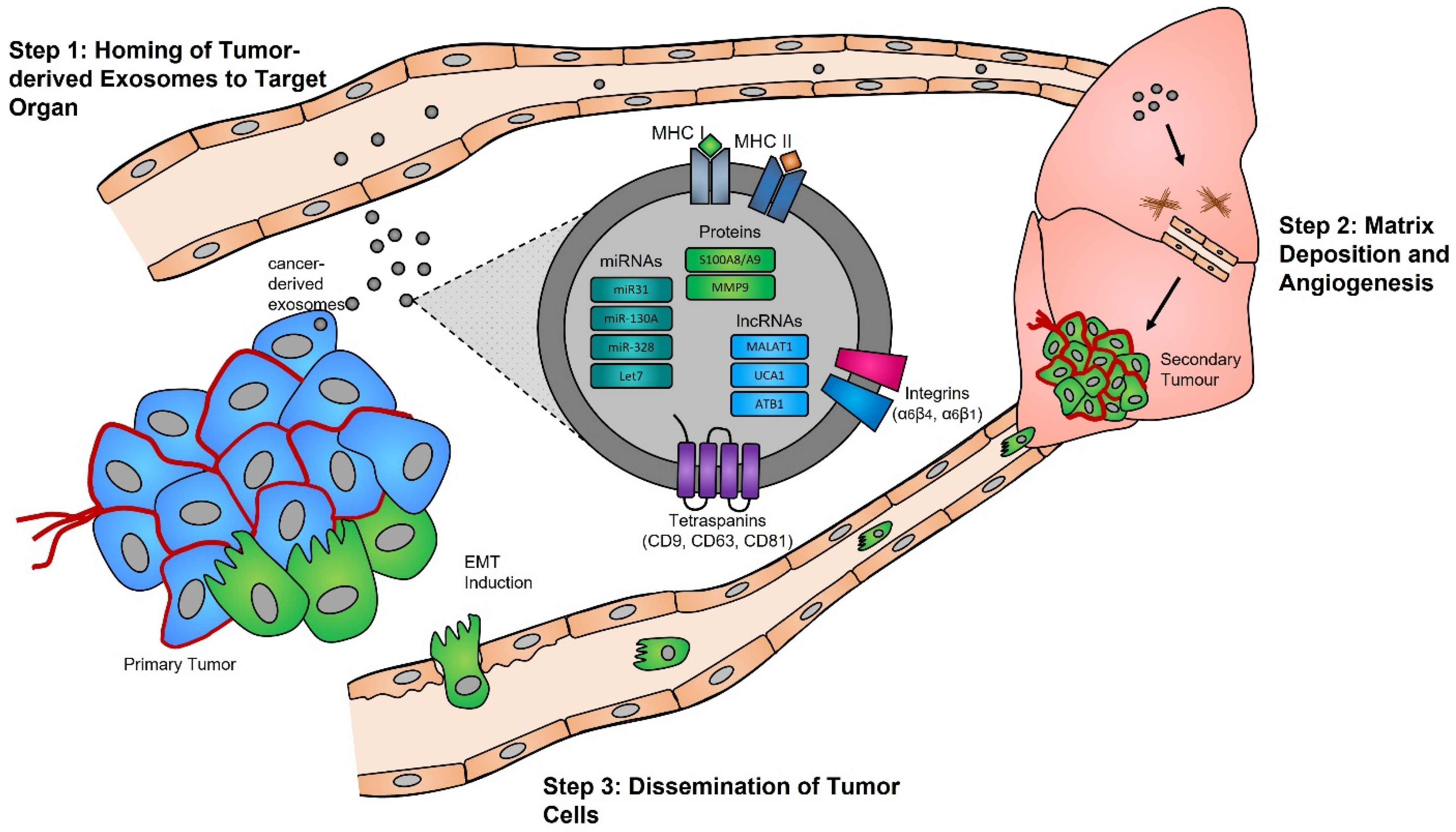
Figure 1. Exosomes and pre-metastatic niche formation. Exosomes released from primary tumors into the circulatory system specifically home to distant target organs (step 1). Upon their arrival, tumor-derived exosomes actively prepare the pre-metastatic niche through myofibroblast activation, induction of angiogenesis, and ECM remodeling (step 2). Local invasion of the primary tumor by cancer cells is followed by their intravasation into the tumor vasculature. These cancer cells survive and travel within the circulatory system, and upon their arrest in capillaries at distant sites, they extravasate into the parenchyma of target organs to commence metastatic colonization (step 3).
An important potential application of circulating tumor exosomes that has been extensively studied is their targeted purification for analysis of their content and identification of tumor biomarkers, including exosome studies of breast cancer [30], lung cancer [31], colon cancer [32][33], and pancreatic cancer [34][35]. Although many options are available for the purification of exosomes from biofluids, very few methods have been proven to provide both the sensitivity and specificity required for detecting low abundance tumor-specific circulating exosomes. Technological advancements aimed at enhancing tissue/cell-specific purification of exosomes from biofluids has significant potential to provide ultra-sensitive molecular assays for detection and monitoring of all human diseases, in particular cancer.
2. Biogenesis and Function of Exosomes in Normal and Pathological Processes
2.1. Biogenesis
Evidence supporting the endosomal origin of exosomes stems from observations that they contain proteins derived exclusively from the cytosol and are devoid of any nuclear proteins [36]. Initial proteomic analyses revealed that while exosomes harbor proteins specific to their cell and tissue of origin, for example integrins (e.g., α6β4, αvβ5, and α6β1 on breast cancer exosomes; αvβ3 and αvβ6 on prostate cancer exosomes; and β4 on pancreatic cancer exosomes), MHC class I and II on immune cell-derived exosomes, as shown for B lymphocytes and dendritic cells [37][38][39], prostate specific antigen (PSA) from prostate cells, asialoglycoprotein receptor 1 (ASGR1) from liver cells [40], microglial proteins (CD11b and CD45) from brain cells [41], placental alkaline phosphatase (PLAP) from placental cells [42][43], and Clara cell protein 16 (CC16) for exosomes produced and released by deep lung cells [44], they also contain proteins common to all exosomes irrespective of their cellular origin, which reflects a highly regulated sorting mechanism [45][46]. For instance, adapter protein ALG-2-interacting protein X (ALIX) and tumor susceptibility gene 101 (TSG101), two proteins extensively characterized for their roles in HIV budding (i.e., release of viral particles from the cell) and MVB formation [47][48], and several tetraspanins (CD63, CD81, CD9, and CD37) are present on all exosomes. The incorporation of specific tetraspanins into exosomes is dependent on cell-type specific needs, as exosomes are involved in a multitude of biological processes, including cell adhesion, motility, invasion, membrane fusion, intracellular signaling, and protein trafficking [49]. To better understand how cancer cells hijack the exosome machinery to regulate the immune response locally, for cell-to-cell communication between neighboring cells, and, as described by Lyden et al. [50], for the successful dissemination of tumor cells, people have divided the endosomal pathway for exosome biogenesis into three major sections (Figure 2).
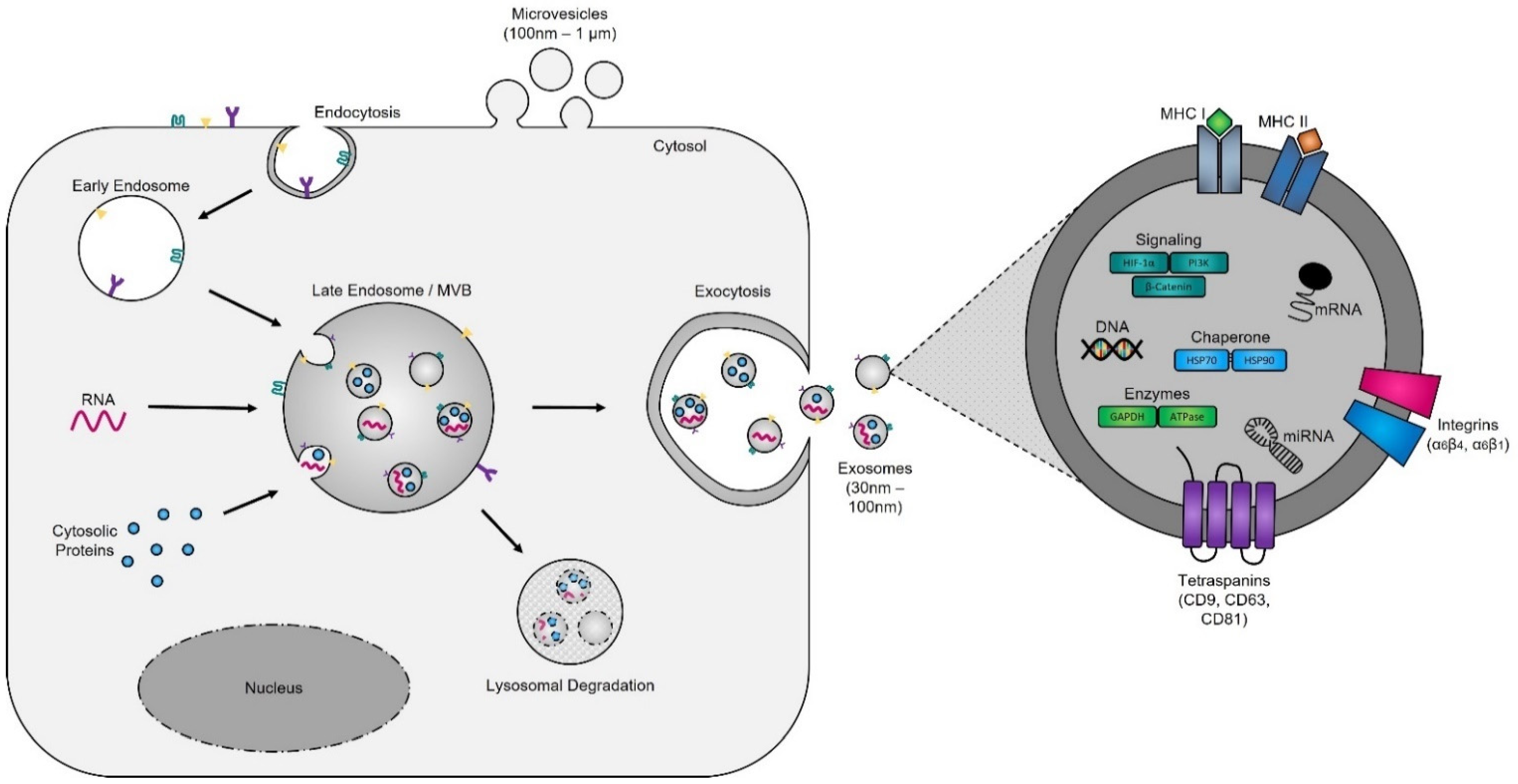
Figure 2. Exosome biogenesis. Exosomes originate from multivesicular bodies (MVBs) (also referred to as late endosomes). The inward budding of the late endosomal membrane around selectively packaged cargo results in the formation of exosomes. The selective packaging of proteins (e.g., tetraspanins, cytoplasmic proteins, and enzymes), nucleic acids (e.g., DNA, RNA, and miRNAs), and lipids (e.g., cholesterol) into exosomes is cell-type dependent and reflects the metabolic status of originating cells. Fusion of MVBs with either lysosomes or the plasma membrane results in either degradation or the release of exosomes into the extracellular matrix, respectively.
2.1.1. The Formation of Early Endosomes
Early endosomes were initially defined as the first cellular compartment to receive incoming endocytic vesicle cargoes [51] but are now recognized to be the main sorting station for initiation of the cellular endocytic pathway [52]. The exact mechanisms underlying the initiation of early endosome formation remain unclear; however, it is thought that their membrane is derived from the fusion of primary endocytic vesicles taken up from the extracellular space [53]. The function of early endosomes (i.e., their role in cargo sorting and delivery of vesicles to the plasma membrane) is heterogenous, cell-type dependent, and defined by the proteins present in their cytosol and on their surface [54]. Individually, early endosomes are complex in structure, with their membrane being comprised of a mosaic of protein subdomains, enriched in Rab5, Rab4, Rab11, Arf1/COPI, retromer, and caveolae of cellular origin [55]. Studies of these proteins have demonstrated that they are responsible for the molecular sorting and the direct transport of endosomes once matured to distinct organelles, including the trans-Golgi network and the plasma membrane [56].
2.1.2. The Maturation of Late Endosomes and Formation of Exosomes
Late endosomes (also termed MVBs) are described as having a multivesicular morphology because they contain intraluminal vesicles (ILVs) [57]. The membrane of these ILVs contains the majority of V-ATPases (i.e., protein pumps responsible for controlling intracellular and extracellular pH), cholesterol, sphingolipid-rich lipid rafts, clathrin coats, and most importantly the endosomal sorting complexes required for transport (ESCRT) machinery [58][59]. This ESCRT machinery sequesters and sorts cytosolic ubiquitinated proteins into ILVs and is the most well-defined and widely understood system for biogenesis of normal and cancer exosomes. The ESCRT complex is comprised of a series of sub-complexes (i.e., ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III) that are sequentially activated by binding of cytosolic ubiquitinated proteins (Figure 3), which are then internalized as early endosomes mature [60].
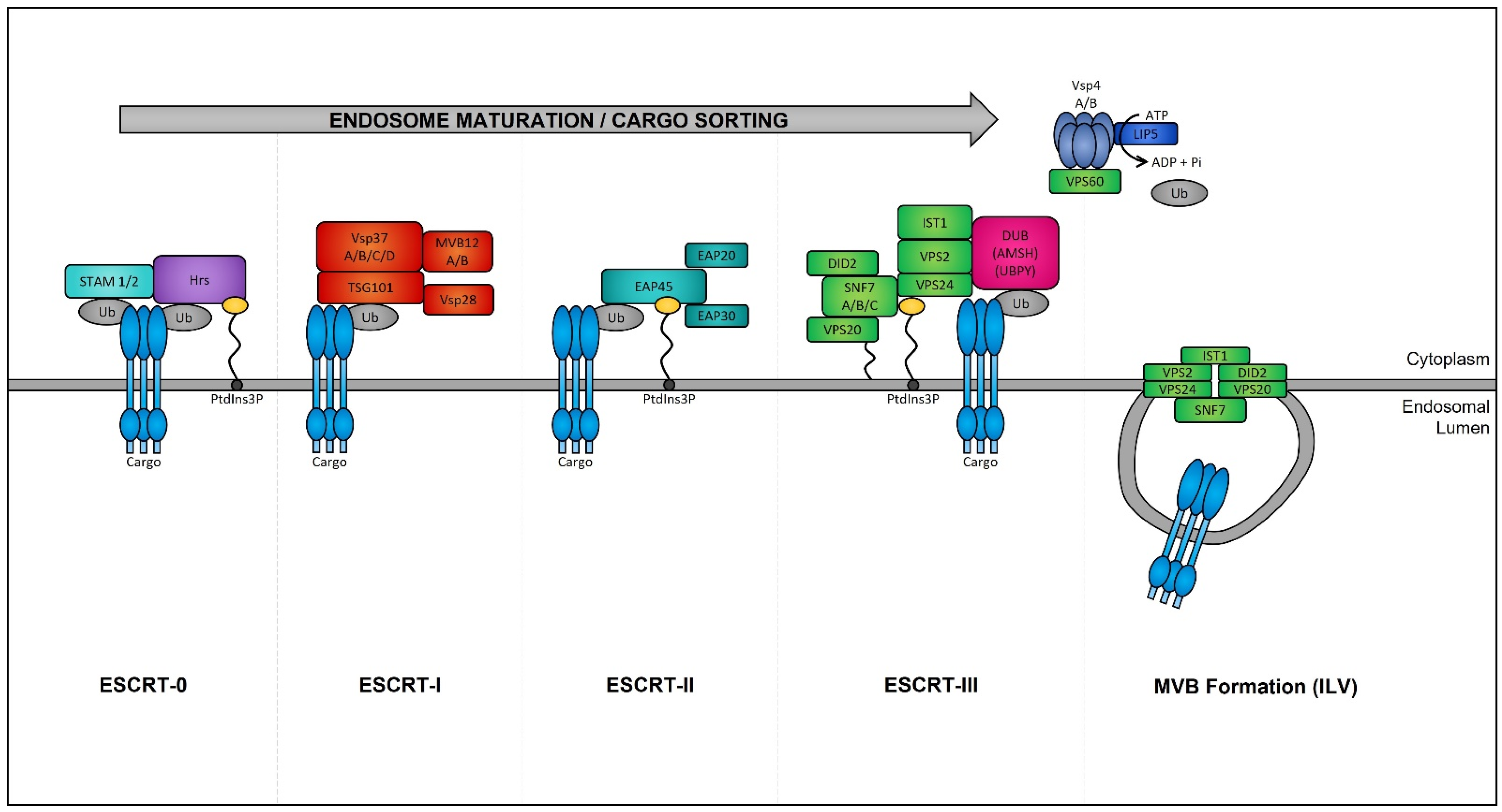
Figure 3. Endosomal sorting complexes required for transport (ESCRT)-dependent MVB formation. ESCRT-dependent MVB formation control the internalization of ubiquitinated proteins into the intraluminal vesicles (ILVs) of MVBs. The ESCRT complex is comprised of a series of sub-complexes which function uniformly during ILV production. ESCRT-0, -I, -II, and -III complexes function consecutively in a stepwise manner to control the selective sorting of ubiquitinated proteins into exosomes.
2.1.3. MVB Trafficking and Exosome Release
The biogenesis of mature MVBs terminates with their progression through one of two pathways: (1) they fuse with lysosomes, resulting in the rapid and irreversible degradation of their contents, or (2) they fuse with the plasma membrane, resulting in the release of their ILV content into the extracellular environment, at which point they are referred to as exosomes [61]. The mechanisms underlying this intracellular switch, which determines the fate of MVBs, is complex and to people's knowledge remains unclear. However, recent studies suggest that both paths are interconnected and that decisions affecting one result in alterations of the other [62]. For instance, two studies showed that inhibition of lysosome fusion with bafilomycin A1 results in the increased secretion of exosomes [63][64]. The release of exosomes by fusion of the MVB with the plasma membrane involves two sequential steps: (i) The targeted movement of MVBs to the plasma membrane, which is reliant on the structure of the microtubule cytoskeleton and the dynamic mechanisms of molecular motors, including dynein, kinesins, and actin-based myosin [65][66], and is regulated by several small Ras-like GTPases (including Rab27 A/B, Rab11, and Rab35), and soluble NSF attachment protein receptor (SNARE) (i.e., small, abundant tail-anchored proteins which are post-translationally inserted into the plasma membrane) proteins [67][68], which have been shown to differ between different cell types [69][70]. (ii) The fusion with the plasma membrane, wherein Rab11 and Rab35 facilitate MVB and plasma membrane fusion [71][72] through the assembly and functionalization of the “SNAREpin” complex. This complex is comprised of one vesicle-SNARE protein (located on MVBs) and three target membrane SNARE proteins (located on the plasma membrane) and is responsible for bringing the two membranes in close apposition resulting in the opening of a fusion pore and the merging of the MVB and plasma membrane [73] and their releasing their exosome cargo into the intercellular space.
2.2. Biological Functions of Exosomes
Exosomes are the most extensively characterized type of extracellular vesicles (typically 30–150 nm in size) [74], which, as described above, are produced via the endosomal pathway (Figure 1) [75]. The release of exosomes is an evolutionary process that has been conserved across all archaea, bacteria, and eukaryotic cells [76]. In humans, exosomes have been shown to regulate cellular functions of neighboring/target cells either by: (1) extracellular interactions of their membrane proteins and lipids with cell-surface receptors and membranes of recipient cells, respectively, to induce signaling cascades [77] or (2) cellular uptake and intracellular pre- or post-transcriptional reprogramming, via the uptake of transcription factors, microRNAs, and long non-coding RNAs, or via the import of foreign messenger RNAs, viral RNAs, and other RNA species [78][79]. Although initially thought to be responsible for the removal of unnecessary material (proteins, lipids, nucleic acids, etc.) [5], exosomes have been shown to play critical roles in a broad range of essential regulatory cellular functions, including the modulation of immunity [80] by regulation of immune cells, particularly by the inhibition of natural killer cells [81] or by acting as antiviral and antibacterial beacons [82][83]. Such activities have also been described with seminal fluid exosomes (originating from seminal glands, prostate, etc.), which protect (i.e., have antimicrobial activities) and provide energy (i.e., produce ATP) to sperm during fertilization [84]. Other functional studies of exosomes have identified their important roles in the maintenance of cellular stemness [85], coagulation of blood and repair of tissues [86], and cardiac and skeletal muscle repair [87][88]. Studies of the function of exosomes highlight the essential roles they play in maintaining the homeostasis of human tissues, which is perturbed during disease [89][90]. Unfortunately, the biological functions of exosomes are “hijacked” in human pathologies, including cardiovascular physiological and pathological disorders (e.g., cardiomyocyte hypertrophy, peripartum cardiomyopathy, and sepsis-induced cardiomyopathy) [91], where exosomes appear to contain erroneous cargoes. Furthermore, exosomes containing aggregation-prone proteins have been found in the cerebral spinal fluid and blood of Parkinson’s disease (PD), Alzheimer’s disease (AD), Creutzfeldt–Jakob disease (CJD), and amyotrophic lateral sclerosis (ALS) patients and may play roles in the propagation of these diseases [92][93][94][95]. Many studies have also highlighted the critical role that tumor exosomes play during the progression of cancer [96]. The best example on the functional diversion of exosomes has been described by the group of David Lyden, which showed that circulating tumor exosomes educate host cells within a secondary organ to prime that environment for establishment of the metastatic niche [97]. In their studies, they demonstrated that exosomes from mouse or human lung, liver, and brain tumor cells preferentially fuse with specific recipient cells (i.e., those at predicted target sites), namely lung epithelial and fibroblast cells, brain endothelial cells, and Kupffer cells in the liver [98].
2.3. Alteration of Exosome Biogenesis in Tumor Cells
Numerous studies have demonstrated that the presence of tumors is associated with a significant increase in the levels of circulating exosomes when compared to healthy individuals [99]. In aggressive brain cancer, glioma cells carrying the EGFRvIII overexpressing mutation exhibit increased secretion of exosomes harboring EGFRvIII [100], and when transferred to astrocytes can confer their oncogenic activity (i.e., ability of exosomes to convey their tumorigenic signals to recipient cells) [101].
ESCRT protein silencing: In order to elucidate the mechanisms underlying the increase in exosome biogenesis observed in cancer cells, Colombo et al. performed a set of iconic experiments to evaluate the role of the different ESCRT components in exosome biogenesis using Hela-CIITA-OVA ovarian cancer cells, which were modified to express MHC-II molecules and allowed for monitoring of exosome secretion [102]. Using this modified cell line, they carried out experiments to silence all twenty-three ESCRT proteins [102]. The silencing of ESCRT-0/I (STAM1, Hrs, and TSG101) resulted in decreased production of CD63, CD81, and MHC-II in exosomes, whereas silencing of ESCRT-III (Vps4B) or ALIX led to increased production and secretion of all exosomes [103]. Three other independent studies, where Hrs (an ESCRT-0 component) was silenced, also led to alterations in exosome secretion [104]. Specifically, the depletion of Hrs in non-cancerous dendritic cells led to a reduction in exosome secretion as measured by the detection of ubiquitinated proteins [104], whereas knockdown of Hrs in HEK293 kidney cancer cells and in head and neck squamous cell carcinoma led to increased exosome secretion and exosomal Wnt3a levels [105]. These studies demonstrated that alterations in ESCRT proteins directly influence exosome production, which may partially help explain their increased production in cancer cells (Figure 4).
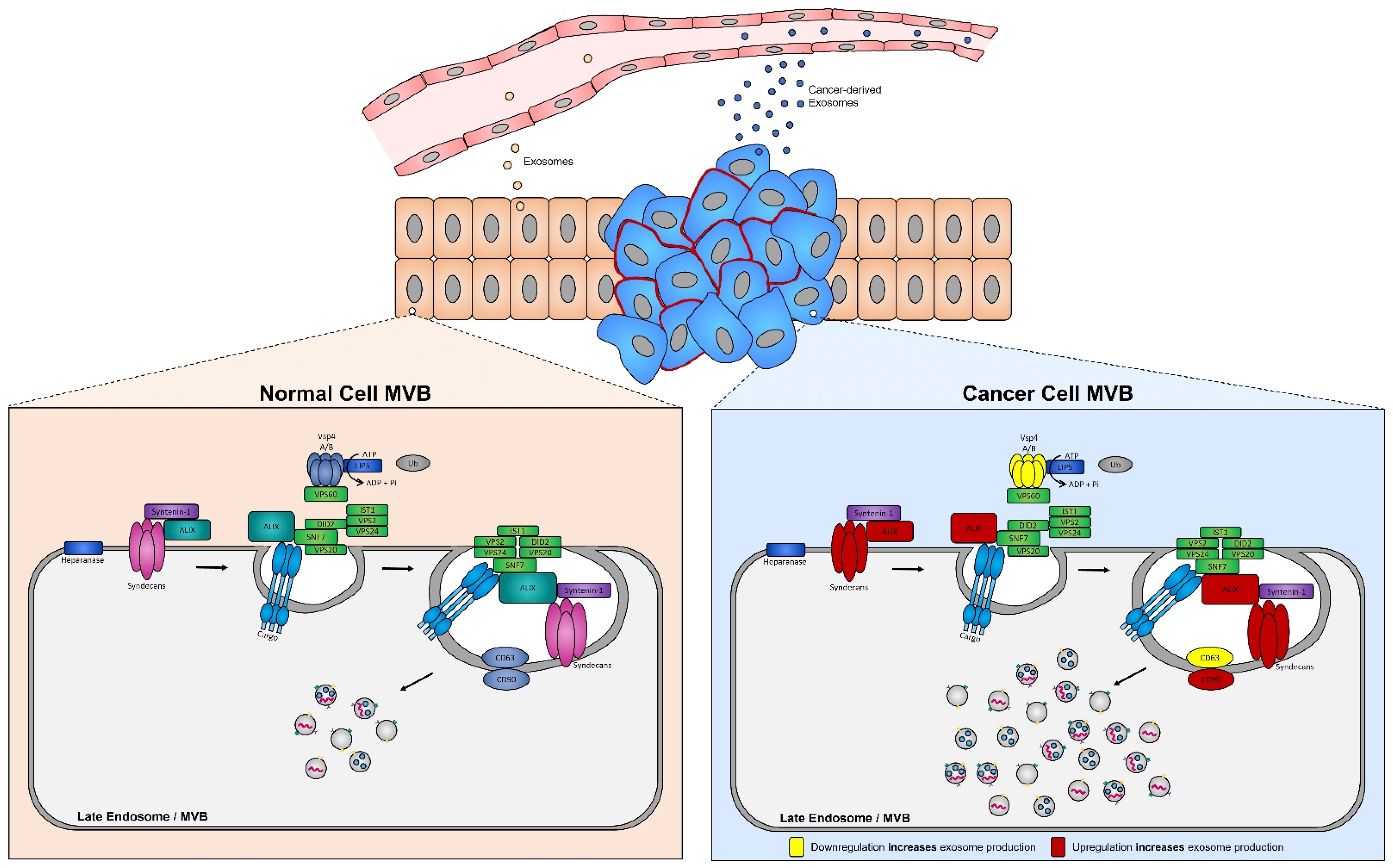
Figure 4. Syndecan–syntenin–ALIX couples to ESCRT-dependent MVB formation in healthy versus cancer cells. Syntenin-1 interacts directly with syndecans and ALIX, the interaction of ALIX with Snf7 of the ESCRT-III complex forms the syndecan–syntenin–ALIX pathway which is directly linked to exosome biogenesis. In cancer, several proteins in this pathway are altered, leading to enhanced exosome production. Alterations leading to the upregulation of either the syndecans, ALIX, and/or CD90 all result in enhanced exosome production, whereas any alterations leading to the downregulation of the ESCRT-III protein Vsp4 A/B and/or CD63 results in the increased production of exosomes seen in cancer cells.
Increased expression of ALIX in tumor cells: The most dramatic alteration in ESCRT-dependent exosome biogenesis for many cancer cell types, including colorectal carcinoma and pancreatic cancer [106][107], has been observed with the overexpression of ALIX, (i.e., induced by a ALIXΔPRR mutation) [108]. ALIX plays an essential role in exosome formation, through stabilization of ESCRT-III, facilitation of ILV formation, and cargo sorting [109]. Thus, upregulation of ALIX expression, as noted in many cancer cell types [110], partially explains the enhanced secretion of exosomes from tumor cells [106].
Increased expression of syntenin-1 in tumor cells: The expression of syntenin-1, which interacts directly with ALIX (via three LYPXnL motifs located within ALIX’s conserved cytoplasmic domains), is upregulated in several cancer types, including breast cancer, lung cancer, and melanoma [111][112][113]. Furthermore, quantitative proteomics identified syntenin-1 to be the most abundant protein present in these exosomes, suggesting that it may serve as a putative cancer biomarker [114]. Syntenin-1 has also been shown to interact with the N-terminus of syndecans (a small family of transmembrane proteoglycans) [115], thus linking them to exosome biogenesis through the syndecan–syntenin–ALIX pathway (i.e., syndecans recruit syntenin-1, which in turn binds to ALIX, and finally ALIX is recruited, stabilizing ESCRT-III and thus increasing exosome biogenesis) [116].
Increased expression of syndecans: Syndecans are a family of four core transmembrane proteins which typically carry two to five heparan sulfate chains [117]. Heparan sulfates are glycosaminoglycans that interact with a wide range of binding proteins, including cytokines, chemokines, growth factors, enzymes, and lipoproteins [118], and are regulated by the heparanase enzyme [119]. Increased expression of heparanase in tumor cells has been associated with increased cell aggressiveness (i.e., invasiveness and metastatic potential) [120]. Because heparanase activity is essential for the regulation of heparan sulfate chains of syndecans, and because syndecans are upregulated in several cancers [121], it is not surprising that heparanase is present in exosomes (ExoCarta, https://www.exoscarta.org/ (accessed on 29 May 2022). Indeed, an increased abundance of heparanase is seen in exosomes isolated from cancer patients compared to those from healthy individuals [122], suggesting enhanced exosomal release via the syndecan–syntenin–ALIX pathway [117][123].
3. Exosomes: A Source of Tumor Biomarkers
Valadi et al. were the first to determine that exosomes carry messenger RNAs (mRNAs) and microRNAs (miRNAs) [17], but since then many studies have demonstrated that exosomes contain a plethora of other RNA species (long-noncoding RNA (lncRNA), piwi RNAs, circular RNAs, etc.) [124]. Exosome whole-transcriptome studies have demonstrated that mRNA molecules are full length and well-protected from RNase activities within circulating exosomes [125]. Some studies have shown that exosomal mRNAs can be transcribed by recipient cells and are possibly used for reprogramming these cells [126]. However, the majority of exosome transcriptomic studies have been performed on microRNAs, because their function as post-transcriptional regulators allows them to control the recipient cells upon delivery [127]. Additional studies on the content of exosomes have also revealed the presence of fragmented genomic DNA, proteins, and lipids from both normal and cancer cells [128]. Fragmented genomic DNA, for example, which is more stable (i.e., protected from degradation) within circulating exosomes than in the circulation, can provide critical biomarkers for the detection of circulating tumor DNA mutations [129]. Castellanos-Rizaldos et al. recently showed that the detection of circulating genomic DNA carrying the EGFRT790M mutation in patients with non-small cell lung cancer (NSCLC) was superior when isolated from circulating exosomes rather than circulating free DNA (cfDNA), particularly in patients with metastatic stage 0/1a lung cancer [130][131]. Overall, several free databases, including Vesiclepedia [132], EVpedia [133], ExoCarta [134] exoRBase [135], and EVmiRNA [136], have been made available for screening the transcriptomes, proteomes and lipidomes of exosomes from different cell types and tissues.
3.1. MicroRNA Biomarkers
MiRNAs represent a large class (~2600 identified in humans) of evolutionarily conserved small regulatory non-coding RNAs expressed from single genes, gene clusters, or intronic passengers, generally 19–25 nucleotides (nt) in length [127]. MiRNAs control gene expression by binding to imperfect complementary sites within the 3′ untranslated regions of their mRNA targets and orchestrate their degradation and/or post-transcriptional repression [137][138]. MiRNAs are involved in the control of all normal biological processes and their expression deregulation (i.e., increased or decreased expression) has been associated with the initiation and development of cancers [139][140][141][142]. Mature and functional cellular miRNAs can be detected in the blood and other biological fluids and can generally be found in two forms: either as cell/membrane-free molecules (i.e., free, bound to Argonaute (Argo) or to nucleophosmin proteins, complexed with high density lipoprotein (HDL)) or encapsulated within extracellular vesicles [124][142][143].
3.1.1. Current Technologies for Quantification of Exosomal MicroRNAs
Since the cargo of exosomes is specifically tailored and to an extent reflects the state and condition of their cells of origin, circulating miRNAs have a potentially strong diagnostic and prognostic value as disease biomarkers [144]. Considering the low amount of material available from exosomes, which may be extracted from limited amounts of biofluids (saliva, sweat, urine, semen, exhaled breath condensates, etc.), current technologies and protocols have been optimized for the global quantification and detection of exosomal miRNAs [145].
High-Throughput Expression Analyses
Next generation sequencing offers a unique unbiased opportunity to globally evaluate the content of exosomes [146][147]. It is important to note that miRNA quantification of cell specific exosomes, either from culture media or biofluids, requires highly purified exosomes [82], because miRNAs are extremely robust [148] and any external contamination can significantly affect quantification. Most next generation miRNA sequencing protocols require relatively high quantities of starting material; for instance, the Illumina TruSeq small-RNA sequencing platform requires a minimum of 10–50 ng of purified small-RNA (Illumina TruSeq small-RNA protocol) or 1000 ng of total RNA [149]. As such, many studies have been focused on global exosomal miRNA purifications from blood or biofluids, which provide workable amounts of total RNA [147][150]. In these studies, exosomes have been rarely isolated from plasma volumes less than 1 mL, and typically these isolations are carried out by means of ultracentrifugation, which yields higher exosome quantities [151]. However, currently many researchers are driving the development towards the utilization of optimized protocols adapted for NGS analysis of low miRNA abundant exosome sub-populations [152]. To this end, one laboratory developed and optimized a cDNA library preparation protocol which allows for 3′ barcoded miRNAs from up to 18 individual samples to be pooled prior to 5′ adapter ligation, reverse transcription, and PCR amplification (~15 to 20 cycles decided via a pilot PCR reaction), which increases the carrier effect of the combined RNA for subsequent enzymatic reactions and purifications [153][154]. This protocol was purposely designed for the analysis of samples with minute quantities of total RNA, and these researchers demonstrated its robustness for exosomal small-RNA analyses with total RNA input <1.5 ng [82]. Using the EV-CATCHER assay for highly pure selection of CD63 exosomes from the serum of patients infected with the SARS-CoV-2 virus and admitted to people's health network, it is able to detect miRNA expression differences in miR-146a and miR-126-3p between COVID-19 patients hospitalized with mild symptoms that did not receive mechanical ventilation and COVID-19 patients hospitalized with severe respiratory symptoms that received mechanical ventilation, within levels estimated at ~25 zeptomoles (10 e-21 mole), which these researchers successfully validated by qPCR [82]. Although the detection miRNAs from low-input total RNA is challenging, their experiments strongly indicate that the specificity and purity of the exosome preparation is critical for the successful detection of unique and rare disease miRNA biomarkers.
Of note, the Nanostring nCounter miRNA platform is an advancing technology for the sensitive, reproducible, and highly multiplexed analysis of up to 800 pre-selected miRNAs. The nCounter miRNA platform is based on a novel method of direct molecular barcoding and digital detection using color-coded probe-pairs [155]. This technology relies on the use of unique miRtags (i.e., oligonucleotide tags), which are ligated to the 3′ end of specific miRNAs. Sequence specificity during ligation is ensured through stepwise controlled annealing and ligation temperature in addition to the use of Tm-optimized bridging oligonucleotides complementary to a portion of the target miRNA and miRtag [156]. To date, a few studies have employed this technology for the quantification of miRNAs from plasma-derived ultracentrifuged exosomes [157]. This methodology is very attractive because it bypasses PCR amplification, which is required during the preparation of small-RNA cDNA libraries, but it may not offer the proper sensitivity yet for the analysis of molecules in the zeptamole concentration range.
Quantitative PCR (qPCR) Analyses
Currently there are numerous commercially available platforms and qPCR-based assays for the analysis of miRNAs (i.e., TaqMan miRNA assays (Thermo Fisher), TaqMan Array microRNA 384-well cards (Thermo Fisher, Tokyo, Japan), OpenArray (Thermo Fisher), SmartChip (Takara, Kusatsu City, Japan), miScript miRNA PCR arrays (Qiagen, Hilden, Germany), miRCURY™ LNA™ Universal RT microRNA PCR (Exiqon, Vedbæk, Denmark), and miRNA Oligo chip (3D Gene)). Quantitative PCR technology is still considered the gold standard for validation of findings made during high-throughput analyses; however, it has inherent biological and experimental limitations for exosomal miRNA studies, which include: (1) The discovery of tumor miRNA panels specific to a tumor exosome remains limited to known or selected miRNAs (which can be multiplexed for up to 700 miRNAs using TaqMan card arrays) rather than global unbiased NGS analyses. In this fashion it does not allow discovery of IsomiR (i.e., miRNA variants of the same miRNA, which can differ in sequence and/or length composition and will bind to different mRNA targets than the original miRNA sequence [158]) expression ratio differences for a specific miRNA in exosome sub-populations. The study of isomiRs is relevant in cancer, as their expression ratio has been shown to be affected by and can elicit changes in tumor cell migration and invasion [159]. (2) qPCR assays typically require large inputs of RNAs for global evaluation and may still require a pre-amplification step (i.e., typically ~14 cycles) before the 40-cycle amplification for fluorescent quantification, which leaves room for potential amplification bias or errors. Most importantly, even when using single qPCR assays, the sensitivity of detection may not be sufficient for miRNA expression validation of total RNA extracted from a selected subpopulation of circulating exosomes. In fact, a group has previously demonstrated that the validation of differentially expressed exosome miRNAs, identified by NGS below 25 zeptomoles, using RT-qPCR is challenging as this quantification lies within the threshold of detection (Ct values of ~36–39) [82]. Studies that have been performed using total RNA extracted from globally circulating exosomes for NSCLC [160], prostate cancer [161], metastatic colorectal cancer [162], and gastric cancer [163] have all failed to provide robust, reproducible, and diagnostic cancer-specific miRNA signatures. The lack of technical standardization and the total RNA purification from globally purified exosomes from plasma (i.e., most of them likely released by blood cells) may have contributed to these limitations.
Droplet Digital PCR (ddPCR) Analysis
Droplet digital PCR (ddPCR) technology has thus become very popular because of its inherent capability for the ultra-sensitive detection of rare molecules [164]. This technology enables absolute quantification by partitioning the reaction into individual droplets (i.e., using water-in-oil and microfluidic technology), wherein molecules are randomly distributed, with each droplet/compartment containing zero, one, or many molecules. In essence this allows for amplification of each compartmentalized PCR mix/target molecule(s) to occur independently of each other [165]. Following amplification, the absorbance of each droplet is counted [166] and using Poisson statistics and the number of positive reactions (i.e., compartments which contain molecules and thus positive signal) the initial copy number and target density are calculated. ddPCR relies on the analysis of individual droplet amplitude and scatter for the absolute quantification of nucleic acids [167]. BioRad has become the leader in ddPCR technology and currently markets their award-winning QX200 AutoDG ddPCR instrument for either EvaGreen or probe-based digital PCR applications. The instrument’s specifications indicate that in each 100 µL volume, up to 20,000 droplets may be generated; generally data from 12,000–16,000 droplets are used for accurate quantification of target molecules (https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf (accessed on 25 May 2022)). Wang et al. recently demonstrated that the analysis of miRNA targets from exosomes (i.e., purified from urine), when compared with traditional TaqMan qPCR technology, was significantly more accurate and sensitive for the detection of low concentrations (i.e., <100 copies/µL) [168]. However, although Bio-Rad offers a large array of mRNA-based detection using their ddPCR instrument, they have not established a standardized protocol for the analysis of miRNAs, and the only available protocols are from published studies [169][170]. The protocol of Hindson et al. suggests that using ddPCR with TaqMan probes improved the day-to-day reproducibility by 7-fold, as seen by a 37–86% decrease in the coefficient of variation, compared to standard qPCR [167]. Considering that tumor exosome miRNA biomarkers may be within or below the zeptomole concentration in a given biofluid, ddPCR technology offers a unique opportunity for the development of ultra-low, -sensitive, and -specific clinical assays and the detection of rare tumor-specific circulating exosomal miRNAs. Standardization of both the tumor exosome isolation techniques (i.e., antigen-based capture, nanoparticle enumeration) and miRNA ddPCR assay preparation will be critical.
3.1.2. Function of MicroRNA in Cancer Exosomes
Exosomal miRNAs from tumor exosomes have been shown to exert a multitude of biological effects during cancer progression which include: (1) the regulation of tumor growth, (2) The evasion from host immune responses, and (3) remodeling of the tumor microenvironment and metastasis.
Regulation of Tumor Growth
Cellular proliferation is a critical aspect to tumor progression, which commonly is a result of the deregulation of cell-cycle related proteins [168], and is a known hallmark of cancer [171]. Research studies have demonstrated that tumor-derived exosomal miRNAs can regulate cancer cell proliferation by targeting cell cycle-associated proteins and signaling proteins [169]. For example, in colorectal cancer, miR-200b is transferred between adjacent cancer cells, where it directly targets 3′-UTRs of p27 and Rho Family GTPase 3 (RND3), two proteins involved in cell cycle regulation [170][172], leading to their downregulation, which in turn induces proliferation in recipient cells [173]. Contrarily, miR-6869-5p, known to act as a tumor suppressor [174], is significantly downregulated in serum-derived exosomes originating from colorectal cancer cells [175]. The downregulation of exosomal miR-6869-5p delivery is associated with enhanced cell proliferation and increased production of the inflammatory cytokines interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) [176]. In metastatic breast cancer, the transfer of exosomal miR-1246 to non-malignant breast cells promotes proliferation, migration, and drug resistance through the direct downregulation of cyclin-G2 (CCNG2) [174]. Moreover, exosomal miRNAs profoundly regulate the apoptotic signaling pathway in cancer cells; for example, Jing et al. demonstrated that the delivery of exosomal miR-769-5p to gastric cancer cells directly targets caspase-9, a cysteine-aspartic protease that serves as the initiator of the intrinsic apoptosis pathway [177], leading to the inhibition of the downstream caspase-dependent apoptotic pathway, and promoting the degradation of the tumor suppressor protein p53 through the ubiquitin–proteosome pathway [178]. Furthermore, in breast cancer, the uptake of exosomal miR-128 leads to a reduction in the expression of the pro-apoptotic protein Bcl-2-associated X protein (Bax) in recipient breast cancer cells [179].
Evasion from Host Immune Responses
Exosomal miRNAs have been shown to function as critical mediators in the crosstalk between cancer cells and macrophages within the tumor microenvironment [180]. Macrophages can be divided into two classes: either M1-polarized or M2-polarized depending on their function [181]. M1 macrophages inhibit tumor growth whereas M2 macrophages have been demonstrated to provide immunosuppression around the tumor niche, while promoting tumor development and metastasis [182]. Exosomes secreted from the SKOV3 epithelial ovarian cancer cell line and containing miR-222-3p can induce macrophage polarization and differentiation to the M2 phenotype by suppression of the cytokine signaling 3 (SOCS3)/STAT3 pathway, which results in tumor growth and enhanced metastatic capability [183]. Similarly, exosomes from hypoxic ovarian cancer cells, present in the center of tumors, deliver miR-940 to macrophages and promote their polarization to the M2 phenotype, which results in tumor progression [184]. In a similar manner, hypoxic glioma-derived exosomes promote M2 macrophage polarization through the delivery of miR-1246, which targets TERF2 interacting protein (TER2IP) via the STAT3 and NF-κB pathways [185]. Dendritic cells are antigen presenting cells (APCs) linking innate and adaptive immunity which express a wide range of toll-like receptors (TLRs) and cytokines [186] that are critical for the activation of host immune responses to pathogens [187], including recognition of cancer cells [188]. In pancreatic cancer, the delivery of exosomes containing miR-203 can inhibit the expression of TLR4 and the production of cytokines, including TNF-α and interleukin-12 (IL-12) in dendritic cells [189], leading to dendritic cell dysfunction, which facilitates cancer cell evasion of the host’s immune response [190]. In addition to the effects that exosomal miRNAs have on macrophages and dendritic cells, they have also been shown to have profound effects on natural killer (NK) cells [191]. NK cells are a part of the innate immune system and can directly kill tumor cells [192] through the release of perforins and granzymes from their cytoplasm [193]. Briand et al. demonstrated that radiotherapy induced the release of exosomes containing increased levels of miR-378a-3p from a variety of cancer cell types, which resulted in the decreased release of granzyme-B from NK cells [194]. Natural killer group 2D receptor (NKG2D) is an activating receptor that is expressed on NK cells, which plays a pivotal role in tumor immunosurveillance [195]. Under hypoxic conditions, the IGR-Heu lung carcinoma and K562 myelogenous leukemia cell lines release exosomes containing increased levels of miR-23a, which when transferred to NK cells results in decreased expression of NKG2D, and thus reduced NK function [196].
Tumor Microenvironment Remodeling and Metastasis
The initiation of metastasis typically occurs when cancer cells undergo a process of epithelial-to-mesenchymal transition (EMT), wherein non-motile epithelial cells transform to a motile/invasive mesenchymal cell type [197]. Once these cells detach from primary tumors and travel through blood vessels, they establish the metastatic niche. Significant evidence suggests that tumor exosomes with their miRNA cargoes play a profound regulatory role in the metastatic process by controlling EMT, angiogenesis (i.e., formation of new blood vessels), niche cell education [198], and tumor cell invasion (Figure 1). Cai et al. demonstrated that exosomes isolated from glioblastoma patients carried increased levels of miR-148a compared to exosomes from healthy volunteers [199]. Exosomal delivery of miR-148a to T98G glioblastoma cells results in cell proliferation and metastasis by downregulation of the cell adhesion molecule 1 (CADM1) and activation of the STAT3 pathway. In triple negative breast cancer, exosomal miR-122 released from breast cancer cells has been shown to reduce the glycolytic pyruvate kinase PKM, and thus decreases glucose uptake in non-tumorigenic cells within the pre-metastatic niche, allowing for increased nutrient availability within the pre-metastatic niche and promoting survival of breast cancer cells that have metastasized [200]. In melanoma, tumor cells exchange miR-222 via exosomes to enhance tumor malignancy by the activation of the phosphoinositide 3-kinase (PI3K)/Akt pathway, via the downregulation of its target gene p27Kip1 [201]. Bone marrow-derived stem cells (BMSCs) present in the hypoxic tumor microenvironment are known to contribute to cancer progression [202]. In lung cancer, hypoxic BMSC-derived exosomes shuttle miR-193a-3p, miR-210-3p, and miR-5100 to lung epithelial cancer cells, which promotes their metastasis via the activation of STAT3 pathway-mediated EMT [203]. Moreover, Yang et al. demonstrated that hepatocarcinoma (HCC) cells with high metastatic potential are able to transfer exosomes containing miR-92a-3p to HCC cells with low metastatic potential, by promoting the EMT-mediated PEN/Akt pathway, as demonstrated by the increased expression of mesenchymal biomarkers (N-cadherin, β-catenin, Snail) and decreased expression of E-cadherin [125]. Altogether, these experiments demonstrate the powerful role that exosomes play in the intercellular communication of cancer cells for the self-propagation of cancer.
Altogether, these studies have demonstrated that cancer cells take advantage of exosomes to transfer tumorigenic miRNAs that can reprogram (i) adjacent tumor cells to promote tumor growth and (ii) a plethora of critical immune cells, to protect the tumor, and (iii) educate pre-metastatic niche cells for the formation of secondary tumor sites before metastasis. However, circulating cancer exosome studies conducted on the same human cancer types have not systematically identified the same deregulated miRNAs, globally suggesting that the miRNA packaging of exosomes may help differentiate healthy individuals from cancer patients (See Table 1). Along with the design of ultra-sensitive and specific cancer exosome purification assays, these studies highlight the potential clinical applications of exosome-based miRNA signatures for non-invasive diagnostic and prognostic evaluation of human cancers.
Table 1. Published miRNA exosome cancer biomarkers identified in human biofluids. Lists of exosomal miRNAs circulating in biofluids of patients diagnosed with colorectal cancer, ovarian cancer, Glioblastoma, liver cancer, pancreatic cancer, lung cancer, extranodal natural killer/T-cell lymphoma, and prostate cancers.
| Cancer Type | Differentially Expressed Between Healthy and Cancer | Reference | |
|---|---|---|---|
| miRNA Biomarkers | |||
| Colorectal Cancer | ↑ | miR-224-5p, miR-548d-5p, miR-200a-3p, miR-320d, miR-200b-3p, miR-1246 | Tang et al., 2019 [162] |
| ↓ | novel_246, novel_301, miR-27a-5p | ||
| miR-135a-5p, miR-204-5p | Sun et al., 2021 [33] | ||
| ↓ | miR-6869-5p | Yan et al., 2018 [174] | |
| ↑ | miR-486-5p, miR-3180-5p | Yan et al., 2017 [175] | |
| ↓ | miR-638, miR-5787, miR-8075, miR-6869-5p, miR-548c-5p | ||
| Ovarian Cancer | ↑ | miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205, miR-214, miR-215 | Taylor and Taylor, 2008 [18] |
| ↑ | miR-940 | Chen et al., 2017 [184] | |
| ↑ | miR-222-3p | Ying et al., 2016 [183] | |
| Glioblastoma | ↑ | let-7a, miR-15b, miR-16, miR-19b, miR-21, miR-26a, miR-27a, miR-92, miR-93, miR-320, miR-20 | Skog et al., 2008 [20] |
| ↑ | miR-148a | Cai et al., 2018 [199] | |
| Liver Cancer | ↑ | miR-17, miR-18a, miR-19a, miR-19b, miR-20a, miR-92a-3p | Yang et al., 2020 [126] |
| ↑ | miR-193a-3p, miR-210-3p, miR-5100 | Zhang et al., 2019 [203] | |
| Pancreatic Cancer | ↑ | miR-21, miR-210 | Wu et al., 2020 [145] |
| ↑ | miR-193a-3p, miR-210-3p, miR-5100 | Zhang et al., 2019 [203] | |
| Lung Cancer | ↑ | miR-132-3p, miR-181b-5p, miR-27a-3p, miR-27b-3p, miR-320a, miR-361-5p, let-7b-5p, miR-24-3p, miR-3184-5p, miR-486-5p, miR-486-3p, miR-320b | Jin et al., 2017 [146] |
| ↓ | let-7a-5p, let-7d-5p, let-7f-5p, miR-26b-5p, miR-30a-3p, miR-30e-3p, miR-744-5p, miR-744-5p, let-7e-5p, miR-191-5p, miR-191-5p, miR-206, miR-21-5p, miR-23a-5p, miR-23b-5p, miR-10b-5p, miR-15b-5p | ||
| miR-30b, miR-30c, miR-103, miR-122, miR-195, miR-203, miR-221, miR-222 | Giallombardo et al., 2016 [160] | ||
| ↑ | miR-193a-3p, miR-210-3p, miR-5100 | Zhang et al., 2019 [203] | |
| Extranodal Natural Killer/T-Cell Lymphoma | ↑ | miR-320e, miR-4454, miR-4516, miR-630, miR-122-5p, miR-574-5p, miR-22-3p, miR-486-3p, miR-1915-5p, miR-1972, miR-1285-5p, miR-222-3p, miR-1305, miR-891b, miR-4455, miR-21-5p, miR-1258, let-7b-5p, miR-25-3p, miR-1268a | Ryu et al., 2020 [157] |
| ↓ | miR-564, miR-196a-5p, miR-520c-3p, let-7d-5p, let-7i-5p, miR-212-3p, miR-29a-3p, miR-608, miR-503-5p, miR-587, miR-548g-3p, miR-765, miR-34c-3p, miR-770-5p, miR-301a-5p, miR-526a, miR-340-5p, miR-325, miR-199a-3p+miR-199b-3p, miR-423-3p | ||
| Prostate Cancer | ↓ | miR-196a-5p, miR-34a-5p, miR-501-3p, miR-92a-1-5p | Rodríguez et al., 2017 [161] |
↑ = miRNA expression observed in circulating exosomes isolated from cancer patients as compared to circulating exosomal miRNAs isolated from healthy individuals and ↓ = decreased miRNA expression observed in circulating exosomes isolated from cancer patients as compared to circulating exosomal miRNAs isolated from healthy individuals.
3.2. Protein Biomarkers
In addition to the many intact RNA species and fragmented genomic DNA available in exosomes, many proteins occupy the surface and interior compartments of these EVs. In the early days, the identification of protein components from EVs was performed by their separation on one-/two-dimensional gels by electrophoresis followed by tandem mass spectrometry [204][205][206][207]. Although these traditional approaches initially allowed for the identification of the mostly abundant proteins, it also allowed for the identification of important exosome protein biomarkers, such as tetraspanins (e.g., CD9, CD63, CD81, and CD151), 14-3-3 proteins, heat shock proteins (e.g., hsp70 and hsp90), and major histocompatibility complex (MHC) [204][205][206][207]. However, during the last two decades, great technical advances in proteomics have been made, which increased sensitivity and have allowed for the identification of thousands of proteins from exosomes and EV proteins obtained from various cell types and body fluids, including serum, urine, saliva, ascites, and breast milk, as shown in several databases (including EVpedia [133], ExoCarta [134], and Vesiclepedia [132]). The ever-growing list of proteins has largely deepened people's understanding of EVs and facilitated the biomedical applications in multiple fields [208][209][210][211][212][213][214][215][216][217].
3.2.1. Protein Extraction from Exosomes and EVs Prior to Analysis
The isolation of exosomes or other EV species is critical to factor proteomic analyses, and it is easy to understand that sample quality directly affects the final proteomics results. Particularly, the excess of certain proteins may overwhelm the proteomic analyses of exosomes as exemplified with the evaluation of exosomes from cell culture media, which often contains a high abundance of albumin due to the addition of serum (i.e., bovine serum), or with exosomes contained in biofluids which carry large amounts of endogenous components (e.g., albumin or other abundant serum proteins from blood samples, uromodulin for urine samples). The presence of such confounding molecules often severely obscures the detection of EV proteins, especially low abundant ones. Therefore, specific procedures should be carefully designed to reduce these non-exosomal proteins or other contaminants, which interfere with proteomic analysis [218][219]. Of note, despite great interest in EVs and their associated proteins, there is currently no consensus on the preferred method(s) for EV isolation. Moreover, it appears that a single isolation method might not reveal the total and specific proteomic content of EVs [220][221]. It is anticipated that the combination of more than one method may be used to describe the proteome content of particular exosome populations, especially considering that exosomes circulating in biofluids represent a heterogeneous population of exosomes from different cellular origins [218][219][220]. Recently a comprehensive and quantitative proteomic analysis was performed on EVs isolated from human primary dendritic cells [222], and it was found that common exosome markers (such as MHC, flotillin, and HSP70) are all identified in all EVs independently of their sedimentation speed, whereas tumor susceptibility 101 (TSG101), syntenin-1, EHD4, Annexin XI, and ADAM10 appeared to be better protein markers for ectosomes than the traditional exosomal protein markers previously used. Undoubtedly, appropriate isolation methods should be adopted for a particular EV population of interest so that meaningful proteomics data can be achieved; the purification methods currently available for selection or purification of exosomes are described in a previous paragraph. As was suggested in this previous section, the combination of different purification methods may help achieve the adequate selection of exosomes from different types of biofluids [218][219][220][221].
3.2.2. Exosome and EV Protein Processing Approaches
Traditional Methods
Unarguably, sample processing is a crucial step for successful proteomics studies.
With traditional proteomics workflows, proteins were often digested in gels or in solutions prior to analyses [204][205][206][207][218][219][220][221]. However, this approach suffers from several inherent drawbacks, such as tedious procedures (i.e., gel digestion and peptide purification/extraction) and lengthy processing time, which limits sample processing throughput and analytical reproducibility.
Filter-Aided Sample Preparation
As an alternative, filter-aided sample preparation (FASP), a simplified method for proteomics sample preparation, has been used for exosome and EV proteomics [223][224][225]. As a relatively new approach, suspension trapping (S-Trap)-based sample preparation has its own advantages [226]. In the S-Trap approach, samples are lysed and solubilized in 5% SDS to create fine particles so that they can be trapped in the filter column. Very strikingly, SDS and other contaminants can be easily removed into the flow-through (even more efficient than the FASP processing). Moreover, proteins retained on the column are readily digested into peptides, without an additional desalting process. All these advantages help further minimize sample loss and increase proteome coverage. Indeed, thorough comparison with the in-solution digestion processing and the FASP method suggests that the S-Trap approach provides the largest number of identifications and the highest reproducibility for EV proteomics [227]. Last but not least, the S-Trap approach allows analysis of substantially lower starting amounts of proteins (down to 50 ~ 200 ng), making it extremely appealing for characterization of minute amounts of EV samples.
3.2.3. Mass Spectrometric Analysis of EV and Exosomal Proteins
Top-Down vs. Bottom-Up Proteomics
Due to the high throughput and its unbiased nature, modern mass spectrometry-based proteomics has allowed for the global identification and quantification of exosomes and other EV proteins [228][229][230][231][232]. Indeed, in the past few decades, quickly evolving proteomics techniques have played an instrumental role in analyzing EV proteins (especially exosomes and ectosomes). As one of the two strategies, top-down proteomics has been tentatively used for the characterization of EV proteins. For example, >200 proteoforms (including multiple proteoforms of the pro-inflammatory mediators S100 A8 and A9) were identified from exosomes shed by murine myeloid-derived suppressor cells [233]. In contrast, bottom-up proteomics has gained much popularity and is widely employed to characterize EV protein cargoes [228][229][230][231][232]. Instead of analyzing intact proteins, proteins are subjected to digestion and then analyzed by tandem mass spectrometry in the bottom-up proteomics strategy. Although almost all major proteomics workflows can be adapted for EV proteomes, challenges remain. Despite great endeavors having been made in the preparation of exosomes from cultured cells or from biofluids (as aforementioned), purification of large amounts of exosomes with high purity is still not routine, which is either a tedious task or inherently limited by the low biofluid volume output. To that end, there is a strong need to develop highly sensitive and robust sample processing procedures and quantification strategy methods so that deep proteomics for EVs can be achieved. Herein, these contents will not summarize all proteomic techniques proposed; rather, these contents will briefly present key factors that determine the output of EV proteomics: the purity/quality of isolated EVs, EV protein sample processing procedures, and tandem mass spectrometry (MS/MS) techniques.
In addition to sample processing, mass spectrometry is another key aspect for EV proteomics. The tandem mass spectrometry-based EV proteomics is usually performed via the data-dependent acquisition (DDA) mode. By combining high-resolution mass spectrometers with isotopic labeling techniques, quantitative information can be obtained for exosome proteomics [228][229][230][231][232]. For example, stable isotopic labelling method using isobaric tags for relative and absolute quantitation (iTRAQ) has been used for urinary EV proteomics and the identification of potential prostate cancer biomarkers, with candidate proteins verified using multiple reaction monitoring (MRM) mass spectrometry (candidate proteins include fatty acid binding protein 5, granulin, AMBP, CHMP4A, and CHMP4C) [234]. Stable isotope labelling with amino acids in cell culture (SILAC) was applied for the proteomics of EVs derived from microsatellite unstable colorectal cancer, allowing for the identification of transforming growth factor beta receptor type 2 (TGFBR2)-regulated EV proteins (such as FN1, GLUL, CTGF, and CDK1) [235]. As a relatively newly emerged approach, label-free data-independent acquisition (DIA) such as SWATH (sequential window acquisition of all theoretical fragment ion) has also been used for the quantitative proteomics of EVs and exosomes [227]. In one study, DDA and SWATH-MS were combined to identify exosome biomarkers indicative of acute or persistent radiation-induced responses [236]. In comparison to total urine proteomics, urine exosome proteomics was demonstrated to be superior for the identification of radiation signatures. Moreover, 23 biomarkers were identified from urine exosomes and 24 biomarkers from serum exosomes post whole-body irradiation (WBI) [236]. Interestingly, proteomic signatures of urinary exosomes seemed to be different from those obtained in serum, with the former indicating injury of the liver, gastrointestinal, and genitourinary track whereas the latter indicated vascular injuries and acute inflammation in response to radiation. Recently, DIA-MS proteomics was used for the analysis of EVs extracted from breast cancer cells, with performance comparison between DDA-MS ad DIA-MS performed [227]. The analyses revealed that DIA-MS outperformed DDA-MS, with deeper EV proteomic coverage and better quantification accuracy achieved [227].
3.2.4. Mass Spectrometric Analysis of EV and Exosomal Protein Modifications
Post-translational modifications (PTMs) add another layer to the regulation of protein functions [237][238]. In comparison to unmodified peptides, modified peptides are present in a low abundance and sub-stoichiometric ratio, making their characterization technically challenging. For such unique and low abundance peptides, enrichment (e.g., TiO2 for phosphopeptides and lectin chromatography for glycopeptides) is necessary prior to mass spectrometry analysis. So far, limited but very appealing studies have been performed for PTM proteomics of exosome samples. For examples, 19 phosphorylation sites corresponding to 14 phosphoproteins [239] and 126 N-glycopeptides belonging to 37 glycoproteins [240] were identified from human urine exosomes. In a recent study, by combining multi-lectin weak affinity chromatography and extensive fractionation (by using high pH reversed-phase chromatography), 378 glycoproteins with 604 glycosylation sites were identified from urine exosomes [241]. By analyzing exosomes isolated from the plasma of breast cancer patients and healthy subjects, a recent phosphoproteomics study identified 9225 and 1014 unique phosphopeptides, with 156 and 271 phosphorylation sites significantly regulated in the microvesicles and exosome fractions, respectively [242]. Among them, three potential phosphopeptide markers belonging to the RALGAPA2, PRKG1, and TJP2 proteins were found to be significantly different between breast cancer patients and healthy subjects. Recently, over 2300 phosphoproteins were identified in breast cancer EVs [243]. Among the identified proteins, many were found to be plasma membrane-based, further indicating that surface protein modifications on cancer exosomes are performed by imparting signaling functions to recipient cells (bypassing the need for the initiation of phosphorylations on membrane receptors). Collectively, although proteomics for PTMs has been largely under-studied in EVs, these very encouraging results suggest that PTMs of proteins should also be of great importance. Comprehensive characterization of protein PTMs in EVs will help understand the sorting mechanisms and identify novel PTM-specific functions of proteins secreted in EVs. They may also help with the discovery of novel biomarkers for translational applications as well as the identification of PTMs with therapeutic effects.
Taken together, proteomics has provided invaluable insights in all aspects of exosome research (i.e., EV biogenesis, secretion, and intercellular interactions). These analyses have revealed that exosomes are enriched with secreted proteins, plasma membrane proteins (e.g., adhesion proteins, receptors, tetraspanins, and transporters), vesicle trafficking-related proteins, cytoskeletal proteins (e.g., actins, myosins, and tubulins), and cytosolic proteins (such as molecular chaperones and metabolic enzymes), which play fundamental roles in their effects on recipient cells as is well studied with cancer metastasis [232][233][234][235][236][243][244]. It is also clear that EV proteomes are dependent not only on the parental cell type and conditions in which the EVs are secreted but are also based on the type of EVs. Very importantly for the diagnostic use of exosomes, the proteome content of each exosome population derived from different sources tends to retain unique signatures and markers of its origin (e.g., specific surface proteins), which may provide features for the early detection of cancer as a minimally invasive liquid biopsy approach.
3.3. Lipid Biomarkers
Lipids are diverse biomolecules that have a plethora of biological functions during developmental, physiological, and pathological processes [245][246][247]. Lipids provide structural integrity to cells and to their subcellular organelles, are an essential part of cellular organization, and allow intracellular trafficking [248][249][250]. Through direct and indirect signaling, lipids can regulate cellular proliferation [251], differentiation [252], and apoptosis [253] but also paly essential roles in energy homeostasis through mitochondrial fatty acid β-oxidation [254]. The dysregulation of lipids in cancer involves alterations of the membrane composition, up/down regulation of pleotropic signaling lipids, and changes in energy metabolism of the transformed cell [255]. These alterations have been linked to cancer cell growth [256], metastasis [257], immune evasion [258] and therapeutic resistance [247]. The study of exosome-derived lipids as cancer biomarkers is an emerging area of research. The lipidomic composition of cell-specific exosomes, their roles in exosome formation and biology, and the methods used for their analysis have been introduced previously and thus will not be discussed in detail here [259][260][261][262][263][264]. A number of eloquent studies have also used lipidomic analysis to identify differences in cancer-derived exosome lipid composition compared to their parental cells [265][266][267][268]. While these studies are fundamental in understanding the composition of exosomes derived from diverse cancer cells, with an aim to elucidating their functional roles, their discussion is out of the scope. This section will focus on the lipid profiles detected from cancer-derived exosomes that have demonstrated utility as diagnostic and prognostic biomarkers, which is a field in its infancy. All lipids presented in the forthcoming sections are annotated according to the nomenclature published by Fahy et al. [269][270].
3.3.1. Methods for the Analysis of Lipids from Exosomes
Mass spectrometry-based technology is the most sensitive and widely used approach for the analysis of lipids from isolated exosomes [271][272][273]. The forthcoming section will provide a brief overview of the different direct and indirect MS methods used for the analysis of lipids from exosomes. Lipids can be analyzed directly from intact exosomes using matrix-assisted laser desorption ionization (MALDI) mass spectrometry [271]. Alternatively, they can be analyzed following one of the four routinely used solvent-based liquid–liquid extraction techniques using both direct and indirect MS methods [272].
Analysis of Intact Exosomes
This technique offers the most rapid and high-throughput analysis of exosomes because they are spotted directly onto the MALDI target without any prior sample preparation. Data acquisition can be carried out within minutes (per sample), providing results that have been shown to match those carried out by MALDI MS of lipid extracts [271]. MALDI is more tolerant to salt impurities when compared to other MS techniques, providing robust and sensitive detection of a wide range of lipid species [274]. A limitation of this technique is the inclusion of high abundant matrix ions that can complicate the resulting mass spectra and the suppression of low abundant lipids or lipids that do not ionize readily. This is particularly evident in positive ion mode, where phosphatidylcholines (PCs) predominate the spectra. There is, however, a number of sample preparation techniques that can be utilized for the targeted analysis of lipids, such as the lithium base-hydrolysis technique introduced recently by Tran et al. which mitigates these ion suppression effects, enabling the sensitive detection of a range of sphingolipids in positive ion mode [275]. Another limitation of both intact analysis and the direct analysis discussed in the following section is the inability of these techniques to separate isomeric and isobaric lipid ions, thereby reducing the number and identification of the lipids detected when compared to indirect methods of analysis. The ability to separate lipid isomers during direct MS analysis of lipids is being realized with the advancement of more sophisticated mass analyzers, including ion mobility and high-resolution platforms, that are enabling the separation of these previously unresolvable lipid ions when analyzed directly [276][277]. Ion mobility platforms employ an electrophoretic-based separation of ions in the gas phase based on their charge, size, and conformation [278]. In this way, ion mobility MS platforms enable the separation of complex lipid mixtures prior to mass analysis [279]. Aside from MALDI, another ion source that has utility for the intact analysis of isolated exosomes is desorption electrospray ionization (DESI). While DESI MS is yet to be utilized for the analysis of exosomes, this technique requires no sample preparation as the samples are placed directly in the ambient ion source whereby desorption and ionization of lipids from the sample are facilitated through a stream of charged solvent (electrospray) that is rapidly applied to the surface of the sample [280]. Thus, DESI MS offers promise as a high-throughput analytical technique of intact exosomes for biomarker investigations.
Direct MS Analysis of Lipid Extracts
The direct analysis of lipid extracts by electrospray ionization (ESI) coupled to tandem MS, without any prior chromatographic separation, was introduced as “shotgun lipidomics” by Han and Gross [281]. Using this technique, lipids are continuously infused into a mass spectrometer and analyzed using simultaneous MS and MS/MS scans, thereby providing structural information that enables their identification and, with the addition of internal standards, enables their quantification [267][282]. The separation of isomeric and isobaric lipids can again be seen as a limitation of this technique; however, with the advancements of the aforementioned instruments and the addition of differential ion mobility to the shotgun lipidomics workflow, the separation of isobars by direct infusion techniques is a now reality [283]. Direct analysis of lipids from exosomes is also possible using MALDI and DESI MS, as discussed for the analysis of lipids from intact exosomes.
Indirect MS Analysis of Lipid Extracts
Indirect analysis of lipids extracted from exosomes involves prior separation utilizing chromatography techniques coupled to MS analysis. Common chromatography techniques include gas chromatography, liquid chromatography, and thin layer chromatography [264][272][284]. The simplest and most cost-effective of these techniques is thin layer chromatography (TLC) coupled to MALDI MS [271]. With this technique, lipid extracts are separated based on their head group polarity using a stationary phase more commonly consisting of silica gel and an apolar solvent mobile phase [285]. The TLC plate is then stained to visualize the lipid bands, which can then either be excised for MALDI MS analysis following an additional lipid extraction protocol or analyzed directly using MALDI imaging [286]. Liquid chromatography-based MS (LC-MS) techniques are the most widely used of the indirect MS methods for lipid analysis [287][288]. As with TLC, the polarity of the mobile and stationary phases is selected based on the selectivity and sensitivity desired [289]. The separation of lipid classes based on their polar head group is more commonly carried out using hydrophilic interaction liquid chromatography (HILIC) coupled to MS [287]. For comprehensive subclass coverage based on lipid fatty acid composition, reverse-phase liquid chromatography (RPLC) is the method of choice [290]. The hydrophobic interactions of RPLC separates lipids based on their carbon chain lengths and levels of saturation, with longer chain polyunsaturated acyl-containing lipids eluting last [287]. The mobile phases used for the elution of lipids can greatly differ from lab to lab, but all are based around organic–aqueous compositions that modify hydrophilic/hydrophobic interactions.
3.3.2. Lipidomic Profiling of Exosomes from Cancer Cells and Cancer Cell Lines
In a study of drug-sensitive vs. drug-resistant non-small lung cell cancer cells, Jung et al. used MALDI-ToF-MS to demonstrate that lipidomic profiles could stratify gefitinib-sensitive from gefitinib-resistant cells based on their exosome composition [291]. They detected 27 lipid signatures that were increased in resistant cells and 40 that were decreased when compared to their drug-sensitive counterpart. Lipids were identified as PCs and ether-linked PCs, lyso-PCs, sphingomyelins (SMs), phosphatidylglycerols (PGs), phosphatidylinositols (PIs), and lyso-PIs with varying fatty acid residues. In a comprehensive metabolic study of the pancreatic cell line PANC-1, the use of UPLC-ESI-Q-TOF-MS allowed identification of diacylglycerol (DAG) (20:2/18:1) as a biomarker of PANC-1 cells undergoing epithelial-to-mesenchymal transition (EMT) [273]. EMT is known to contribute to both metastatic disease and therapeutic resistance of pancreatic cancer cells [292][293]. These two studies demonstrate the utility of lipidomic exosome profiling for biomarker discovery, with potential to drive precision medicine by prediction of treatment response. LC-MS/MS analysis of EVs isolated from the prostate cell line, RWPE1, the prostate cancer cell line, NB26, and the metastatic/hormone resistant prostate cancer cell line, PC-3, showed similar lipid profiles when the mean abundance of lipid classes was compared [294]. Analysis of lipid subclasses and their fatty acyl residue composition from each sample, however, enabled the stratification of normal, cancer, and metastatic prostate cancer cells based on specific lipid profiles. In total, 187 lipid species were quantified as showing a differential abundance between these cell lines, highlighting the complexity of lipid analysis when considering full structural composition of each lipid class. DAG and triacylglycerol (TAG) species were decreased in tumor and metastatic cells when compared to their normal counterpart, with DG (16:0/22:6) detected with the highest fold change difference between normal vs. prostate cancer cells lines. Conversely, a number of PCs, SMs, phosphatidylethanolamine (PEs), ceramides (Cers), hexosylceramides (HexCers), phosphatidylserine (PS), PIs, cholesteryl esters (CEs), and ether-phospholipids (PLs), were enriched in both tumor and metastatic cell lines to degrees that were dependent on the structural combination of their subclass (head group) and fatty acid residues. A panel of biomarkers that could stratify prostate cancer and prostate cancer vs. metastatic disease was not proposed, possibly due to the number of lipids detected and the processing method utilized to handle such complex data. Another extensive lipidomic study comparing the lipidomic profiles of exosomes from the ovarian cancer cell line, SKOV-3, to those detected in exosomes from normal ovarian surface epithelial cell line, HOSEPiC, identified 110 significantly altered lipids from over 1200 identified [295]. These again encompassed the phospholipid, sphingolipid, and sterol classes already mentioned in previous studies and these differences were dependent on the fatty acyl composition of the subclasses detected. The authors highlighted PG (34:1) and CE (18:2) as the lipids with the highest significance between the cell lines, along with PS (36:2) as a SKOV-3 cancer-specific EV lipid, and proposed the utility of these as potential biomarkers. Lipidomic analysis of EVs secreted from pancreatic ductal adenocarcinoma cells (PDACs) allowed detection of increased numbers of the pleotropic signaling lipid, ceramide-1-phosphate (C1P) [296]. While this was a functional study demonstrating that EVs containing C1P can promote pancreatic stem cell motility, the specificity of this sphingolipid subclass along with the report of C1P (d18:1/16:0) being the predominant species detected warrants further investigation for its use as a PDAC-specific EV biomarker.
3.3.3. Lipidomic Profiling of Exosomes from Liquid Biopsies
Lipidomic analysis of exosomes isolated from patient plasma samples successfully classified early and late-stage non-small cell lung cancer (NSCLC) by using the 430 detectable lipid ions [297]. Using ultra-high resolution mass spectrometry without any prior chromatography separation, Fan et al. were able to discriminate the different stages of NSCLC vs. healthy individuals from 91 patient samples. The data were classified using two different multivariate statistical analysis methods, random forests (RF) and least absolute shrinkage and selection operator (LASSO) regression analysis. RF analysis identified a higher number of lipid signatures with discriminating capabilities, of which 15 were identified and validated as PC (18:1/18:2), PC (18:0/18:2), PC (16:0/22:6), PC (16:0/18:2), SM (d18:1/16:0), PC (18:0/20:3), PC (16:0/20:4), PC (16:0/22:5), CE (20:4), TAG (52:5), SM (d18:1/24:1), PC (18:0/18:1), PC (16:0/16:0), TAG (54:6), and LysoPC (16:0). Of the seven LASSO features detected with discriminatory capabilities, three were identified and validated and they overlapped with those detected during the RF analysis, which increased the power of these lipids as classifiers for diagnosing and stratifying NSCLC. The three overlapping species were PC (18:1/18:2), PC (18:0/20:3), and TAG (54:6). Their study showed that despite the discriminatory area under the ROC (aucROC) curve values being higher for RF analysis, the specificity was higher when using LASSO and fewer lipid species. Their study demonstrates the power of a relatively small number of lipid species as valid diagnostic and prognostic biomarkers for cancer detection. The analysis of lipid profiles from serum exosomes isolated from pancreatic cancer (PC) patients identified 20 lipid species that were able to discriminate PC from healthy controls [298]. By correlating the data on the dysregulated lipids with clinical data, this group was able to identify several lipid species that provided a prognostic value. LysoPC (22:0) was shown to correlate with tumor stage, diameter, lymphocyte count, and the diagnostic markers CA19-9 and CA242. PC (P-14:0/22:2) was also shown to correlate with tumor stage and lymphocyte count, but this lipid species also correlated with three diagnostic markers, CA19-9, CA242, and CEA. PE (16:0/18:1) was also associated with tumor stage and the CA19-9 and CA242 diagnostic markers. This lipid, however, was shown to be an independent prognostic factor that correlated with patients’ overall survival. In a study comparing the lipidomic profiles of exosomes isolated from urine samples taken from prostate cancer patients compared to healthy controls, Skotland et al. identified nine lipid species that were significantly different between the two groups [299]. HexCer (d18:1/16:0), LacCer (d18:1/16:0), and PC (16:0/18:1) were all found to increase in cancer exosomes relative to the control. Contrarily, PE (O-18:0/18:1), PE(P-36:4/O-36:5), PE (P-18:1/20:4), PS (16:0/18:1), PS (18:0/18:1), and PS (18:1/18:1) were found to decrease in cancer exosomes relative to the controls. Despite the statistical differences of these lipids between prostate cancer and healthy patient urinary exosomes, their diagnostic performance was poor when used alone, with sensitivity values ranging from 20 to 67%. However, when the authors processed their data taking into account lipid ratio comparisons, the ratios of LacCer (d18:1/16:0) to PS (18:1/18:1) and PS (18:0/18:2) significantly separated the prostate cancer from healthy controls with a sensitivity of 93% and a specificity of 100%. Their study again provides evidence that a very small number of lipid species are capable of acting as diagnostic exosome-based biomarkers, but also that care needs to be taken during data analysis and interpretation to ensure rigor in the identification of lipid-based biomarker panels from liquid biopsies.
3.3.4. Lipidomic-Specific Exosome Isolation for Use as Cancer Biomarkers
A number of studies have reported the utility of exosomes containing PS lipid species as diagnostic biomarkers. Although not technically lipidomic profiling analysis per se, PS-protein binding affinity assays have been used for the capture of cancer-based exosomes from cell culture and biological fluids [300][301][302][303][304]. In normal healthy cells, PS lipids are predominantly located on the inner leaflet of the lipid membrane, but in a diverse number of cancer cells, PS has been shown to externalize to the outer leaflet, and this has garnered interest for use as a novel biomarker and therapeutic target [305][306][307][308]. In a similar regard, tumor cell-derived exosomes also contain increased numbers of PS species that are predominantly located in the external membrane bilayer, where they are believed to play a role in the uptake of cancer-based exosomes by recipient cells [260][263][309]. In a proof-of-concept study, Lea et al. showed that PS-expressing exosomes detected from patient plasma could be used as a diagnostic biomarker for ovarian malignancies [310]. The group developed a stringent ELISA for selective binding and detection of PS-expressing exosomes that distinguished healthy individuals from patients with ovarian malignancies. The group also demonstrated the utility of their assay in detecting tumor recurrence from plasma-based liquid biopsies. Their study is a pivotal study as there are currently no standard diagnostic or prognostic biomarkers for ovarian cancers, and grading is often carried out following surgical resection and histological staining/analysis. The same group also utilized the PS ELISA-based assay system to quantify picogram amounts of PS-expressing exosomes from the blood of tumor-bearing mice [302]. In four preclinical model systems, the PS-expressing exosomes were detected weeks prior to the development of tumor masses, indicating that circulating PS-expressing exosomes could provide an excellent source of biomarkers for the early diagnosis of a number of tumors.
In summary, while the field of exosome-derived lipids as cancer biomarkers is in its infancy, the aforementioned studies demonstrate their promising utility as diagnostic and prognostic markers. The advancement of sophisticated MS instrumentation for the sensitive and high-throughput analysis of lipids will enable the rapid translation of data from the bench to the clinic. The biggest caveat is standardizing both MS workflows and the analysis of the complex lipid data from these studies. Standardized workflows, along with validated biomarker panels, are essential to enable the incorporation of exosome-derived lipids into clinical diagnostics.
This entry is adapted from the peer-reviewed paper 10.3390/cancers14143350
References
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197.
- Hargett, L.A.; Bauer, N.N. On the origin of microparticles: From “platelet dust” to mediators of intercellular communication. Pulm. Circ. 2013, 3, 329–340.
- Chargaff, E. Cell structure and the problem of blood coagulation. J. Biol. Chem. 1945, 160, 351–359.
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339.
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978.
- Johnstone, R.M.; Mathew, A.; Mason, A.B.; Teng, K. Exosome formation during maturation of mammalian and avian reticulocytes: Evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell Physiol. 1991, 147, 27–36.
- Gawrisch, K.; Stibenz, D.; Möps, A.; Arnold, K.; Linss, W.; Halbhuber, K.J. The rate of lateral diffusion of phospholipids in erythrocyte microvesicles. Biochim. Biophys. Acta 1986, 856, 443–447.
- Vidal, M.; Sainte-Marie, J.; Philippot, J.R.; Bienvenue, A. Asymmetric distribution of phospholipids in the membrane of vesicles released during in vitro maturation of guinea pig reticulocytes: Evidence precluding a role for “aminophospholipid translocase”. J. Cell Physiol. 1989, 140, 455–462.
- Vidal, M.J.; Stahl, P.D. The small GTP-binding proteins Rab4 and ARF are associated with released exosomes during reticulocyte maturation. Eur. J. Cell Biol. 1993, 60, 261–267.
- Lee, Y.J.; Jy, W.; Horstman, L.L.; Janania, J.; Reyes, Y.; Kelley, R.E.; Ahn, Y.S. Elevated platelet microparticles in transient ischemic attacks, lacunar infarcts, and multiinfarct dementias. Thromb. Res. 1993, 72, 295–304.
- Singh, N.; Gemmell, C.H.; Daly, P.A.; Yeo, E.L. Elevated platelet-derived microparticle levels during unstable angina. Can. J. Cardiol. 1995, 11, 1015–1021.
- Powell, J.J.; Harvey, R.S.; Thompson, R.P. Microparticles in Crohn’s disease—Has the dust settled? Gut 1996, 39, 340–341.
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.-S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013, 13, 3354–3364.
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.W.; Geuze, H.J.; Stoorvogel, W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003, 278, 10963–10972.
- Bard, M.P.; Hegmans, J.P.; Hemmes, A.; Luider, T.M.; Willemsen, R.; Severijnen, L.A.; van Meerbeeck, J.P.; Burgers, S.A.; Hoogsteden, H.C.; Lambrecht, B.N. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol. 2004, 31, 114–121.
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212.
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673.
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21, Erratum in Gynecol. Oncol. 2010, 116, 153.
- Rosell, R.; Wei, J.; Taron, M. Circulating MicroRNA Signatures of Tumor-Derived Exosomes for Early Diagnosis of Non-Small-Cell Lung Cancer. Clin. Lung Cancer 2009, 10, 8–9.
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476.
- Janowska-Wieczorek, A.; Wysoczynski, M.; Kijowski, J.; Marquez-Curtis, L.; Machalinski, B.; Ratajczak, J.; Ratajczak, M.Z. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 2005, 113, 752–760.
- Hao, S.; Ye, Z.; Li, F.; Meng, Q.; Qureshi, M.; Yang, J.; Xiang, J. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp. Oncol. 2006, 28, 126–131.
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092.
- Iguchi, H.; Kosaka, N.; Ochiya, T. Secretory microRNAs as a versatile communication tool. Commun. Integr. Biol. 2010, 3, 478–481.
- Jung, T.; Castellana, D.; Klingbeil, P.; Cuesta Hernández, I.; Vitacolonna, M.; Orlicky, D.J.; Roffler, S.R.; Brodt, P.; Zöller, M. CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia 2009, 11, 1093–1105.
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell Proteom. 2010, 9, 1085–1099.
- Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 1889, 133, 571–573.
- Mathias, R.A.; Gopal, S.K.; Simpson, R.J. Contribution of cells undergoing epithelial-mesenchymal transition to the tumour microenvironment. J. Proteom. 2013, 78, 545–557.
- Liu, C.M.; Hsieh, C.L.; Shen, C.N.; Lin, C.C.; Shigemura, K.; Sung, S.Y. Exosomes from the tumor microenvironment as reciprocal regulators that enhance prostate cancer progression. Int. J. Urol. 2016, 23, 734–744.
- Halvaei, S.; Daryani, S.; Eslami, S.Z.; Samadi, T.; Jafarbeik-Iravani, N.; Bakhshayesh, T.O.; Majidzadeh, A.K.; Esmaeili, R. Exosomes in Cancer Liquid Biopsy: A Focus on Breast Cancer. Mol. Ther. Nucleic Acids 2018, 10, 131–141.
- Chen, R.; Xu, X.; Qian, Z.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. The biological functions and clinical applications of exosomes in lung cancer. Cell Mol. Life Sci. 2019, 76, 4613–4633.
- Jia, S.; Zhang, R.; Li, Z.; Li, J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget 2017, 8, 55632–55645.
- Sun, B.; Li, Y.; Zhou, Y.; Ng, T.K.; Zhao, C.; Gan, Q.; Gu, X.; Xiang, J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J. Cell Physiol. 2019, 234, 1416–1425.
- Lu, L.; Risch, H.A. Exosomes: Potential for early detection in pancreatic cancer. Future Oncol. 2016, 12, 1081–1090.
- Chen, K.; Wang, Q.; Kornmann, M.; Tian, X.; Yang, Y. The Role of Exosomes in Pancreatic Cancer from Bench to Clinical Application: An Updated Review. Front. Oncol. 2021, 11, 644358.
- Morita, E.; Sandrin, V.; Chung, H.-Y.; Morham, S.G.; Gygi, S.P.; Rodesch, C.K.; Sundquist, W.I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007, 26, 4215–4227.
- Théry, C.; Duban, L.; Segura, E.; Véron, P.; Lantz, O.; Amigorena, S. Indirect activation of naïve CD4+ T cells by dendritic cell-derived exosomes. Nat. Immunol. 2002, 3, 1156–1162.
- Buschow, S.I.; Nolte-‘t Hoen, E.N.; van Niel, G.; Pols, M.S.; Ten Broeke, T.; Lauwen, M.; Ossendorp, F.; Melief, C.J.; Raposo, G.; Wubbolts, R.; et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 2009, 10, 1528–1542.
- Buschow, S.I.; van Balkom, B.W.; Aalberts, M.; Heck, A.J.; Wauben, M.; Stoorvogel, W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 2010, 88, 851–856.
- Newman, L.A.; Useckaite, Z.; Johnson, J.; Sorich, M.J.; Hopkins, A.M.; Rowland, A. Selective Isolation of Liver-Derived Extracellular Vesicles Redefines Performance of miRNA Biomarkers for Non-Alcoholic Fatty Liver Disease. Biomedicines 2022, 10, 195.
- Chiasserini, D.; van Weering, J.R.; Piersma, S.R.; Pham, T.V.; Malekzadeh, A.; Teunissen, C.E.; de Wit, H.; Jiménez, C.R. Proteomic analysis of cerebrospinal fluid extracellular vesicles: A comprehensive dataset. J. Proteom. 2014, 106, 191–204.
- Pillay, P.; Maharaj, N.; Moodley, J.; Mackraj, I. Placental exosomes and pre-eclampsia: Maternal circulating levels in normal pregnancies and, early and late onset pre-eclamptic pregnancies. Placenta 2016, 46, 18–25.
- Miranda, J.; Paules, C.; Nair, S.; Lai, A.; Palma, C.; Scholz-Romero, K.; Rice, G.E.; Gratacos, E.; Crispi, F.; Salomon, C. Placental exosomes profile in maternal and fetal circulation in intrauterine growth restriction—Liquid biopsies to monitoring fetal growth. Placenta 2018, 64, 34–43.
- Seibold, T.; Schönfelder, J.; Weeber, F.; Lechel, A.; Armacki, M.; Waldenmaier, M.; Wille, C.; Palmer, A.; Halbgebauer, R.; Karasu, E.; et al. Small Extracellular Vesicles Propagate the Inflammatory Response after Trauma. Adv. Sci. 2021, 8, e2102381.
- Larssen, P.; Wik, L.; Czarnewski, P.; Eldh, M.; Löf, L.; Ronquist, K.G.; Dubois, L.; Freyhult, E.; Gallant, C.J.; Oelrich, J.; et al. Tracing Cellular Origin of Human Exosomes Using Multiplex Proximity Extension Assays. Mol. Cell Proteom. 2017, 16, 1547, Erratum in Mol. Cell. Proteom. 2017, 16, 502–511.
- Valadi, H.; Eksröm, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Bissig, C.; Gruenberg, J. ALIX and the multivesicular endosome: ALIX in wonderland. Trends Cell Biol. 2014, 24, 19–25.
- Alenquer, M.; Amorim, M.J. Exosome biogenesis, regulation, and function in viral infection. Viruses 2015, 7, 5066–5083.
- Hemler, M.E. Targeting of tetraspanin proteins—Potential benefits and strategies. Nat. Rev. Drug Discov. 2008, 7, 747–758.
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360.
- Helenius, A.; Mellman, I.; Wall, D.; Hubbard, A. Endosomes. Trends Biochem. Sci. 1983, 8, 245–250.
- Elkin, S.R.; Lakoduk, A.M.; Schmid, S.L. Endocytic pathways and endosomal trafficking: A primer. Wien. Med. Wochenschr. 2016, 166, 196–204.
- Alberts, B.; Johnson, A.; Lewis, J. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; Transport into the Cell from the Plasma Membrane: Endocytosis. Available online: https://www.ncbi.nlm.nih.gov/books/NBK26870/ (accessed on 26 May 2022).
- Kaur, G.; Lakkaraju, A. Early Endosome Morphology in Health and Disease. Adv. Exp. Med. Biol. 2018, 1074, 335–343.
- Hotaru, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500.
- Pfeffer, S.R. Multiple routes of protein transport from endosomes to the trans Golgi network. FEBS Lett. 2009, 583, 3811–3816.
- Piper, R.C.; Katzmann, D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547.
- Lu, A.; Tebar, F.; Alvarez-Moya, B.; López-Alcalá, C.; Calvo, M.; Enrich, C.; Agell, N.; Nakamura, T.; Matsuda, M.; Bachs, O. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J. Cell Biol. 2009, 184, 863–879.
- Frankel, E.B.; Audhya, A. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol. 2018, 74, 4–10.
- Katzmann, D.J.; Babst, M.; Emr, S.D. Ubiquitin-Dependent Sorting into the Multivesicular Body Pathway Requires the Function of a Conserved Endosomal Protein Sorting Complex, ESCRT-I. Cell 2001, 106, 145–155.
- Piper, R.C.; Luzio, J.P. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr. Opin. Cell Biol. 2008, 19, 459–465.
- Peng, X.; Yang, L.; Ma, Y.; Li, Y.; Li, H. Focus on the morphogenesis, fate and the role in tumor progression of multivesicular bodies. Cell Commun. Signal. 2020, 18, 122.
- Alvarez-Erviti, L.; Seow, Y.; Schapira, A.H.; Gardiner, C.; Sargent, I.L.; Wood, M.J.; Cooper, J.M. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 2011, 42, 360–367.
- Miao, Y.; Li, G.; Zhang, X.; Xu, H.; Abraham, S.N. A TRP Channel Senses Lysosome Neutralization by Pathogens to Trigger Their Expulsion. Cell 2015, 161, 1306–1319.
- Yoon, S.; Kovalenko, A.; Bogdanov, K.; Wallach, D. MLKL, the Protein that Mediates Necroptosis, Also Regulates Endosomal Trafficking and Extracellular Vesicle Generation. Immunity 2017, 47, 51–65.e7.
- Olenick, M.A.; Holzbaur, E.L.F. Dynein activators and adaptors at a glance. J. Cell Sci. 2019, 132, jcs227132.
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011, 91, 119–149.
- Koike, S.; Jahn, R. SNARE proteins: Zip codes in vesicle targeting? Biochem. J. 2022, 479, 273–288.
- Lorentz, A.; Baumann, A.; Vitte, J.; Blank, U. The SNARE Machinery in Mast Cell Secretion. Front. Immunol. 2012, 3, 143.
- Homma, Y.; Hiragi, S.; Fukuda, M. Rab family of small GTPases: An updated view on their regulation and functions. FEBS J. 2021, 288, 36–55.
- Yu, X.; Prekeris, R.; Gould, G.W. Role of endosomal Rab GTPases in cytokinesis. Eur. J. Cell Biol. 2007, 86, 25–35.
- Ramel, D.; Wang, X.; Laflamme, C.; Montell, D.J.; Emery, G. Rab11 regulates cell-cell communication during collective cell movements. Nat. Cell Biol. 2013, 15, 317–324.
- Heo, P.; Coleman, J.; Fleury, J.B.; Rothman, J.E.; Pincet, F. Nascent fusion pore opening monitored at single-SNAREpin resolution. Proc. Natl. Acad. Sci. USA 2021, 118, e2024922118.
- Mathieu, M.; Névo, N.; Jouve, M.; Valenzuela, J.I.; Maurin, M.; Verweij, F.J.; Palmulli, R.; Lankar, D.; Dingli, F.; Loew, D.; et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 2021, 12, 4389.
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11.
- Gerlach, P.; Schuller, J.M.; Bonneau, F.; Basquin, J.; Reichelt, P.; Falk, S.; Conti, E. Distinct and evolutionary conserved structural features of the human nuclear exosome complex. Elife 2018, 7, e38686.
- Bahrami, A.; Moradi Binabaj, M.; AFerns, G. Exosomes: Emerging modulators of signal transduction in colorectal cancer from molecular understanding to clinical application. Biomed. Pharmacother. 2021, 141, 111882.
- Schwarzenbach, H.; Gahan, P.B. MicroRNA Shuttle from Cell-To-Cell by Exosomes and Its Impact in Cancer. Noncoding RNA 2019, 5, 28.
- Liu, H.; Chen, W.; Zhi, X.; Chen, E.J.; Wei, T.; Zhang, J.; Shen, J.; Hu, L.Q.; Zhao, B.; Feng, X.H.; et al. Tumor-derived exosomes promote tumor self-seeding in hepatocellular carcinoma by transferring miRNA-25-5p to enhance cell motility. Oncogene 2018, 37, 4964–4978.
- Iguchi, Y.; Eid, L.; Parent, M.; Soucy, G.; Bareil, C.; Riku, Y.; Kawai, K.; Takagi, S.; Yoshida, M.; Katsuno, M.; et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 2016, 139 Pt 12, 3187–3201.
- Vulpis, E.; Soriani, A.; Cerboni, C.; Santoni, A.; Zingoni, A. Cancer Exosomes as Conveyors of Stress-Induced Molecules: New Players in the Modulation of NK Cell Response. Int. J. Mol. Sci. 2019, 20, 611.
- Mitchell, M.I.; Ben-Dov, I.Z.; Liu, C.; Ye, K.; Chow, K.; Kramer, Y.; Gangadharan, A.; Park, S.; Fitzgerald, S.; Ramnauth, A.; et al. Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): A customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating small-EVs. J. Extracell. Vesicles 2021, 10, e12110.
- Troyer, Z.; Tilton, J.C. Extracellular vesicles as carriers of viruses. ExRNA 2021, 3, 13.
- Göran Ronquist, K. Extracellular vesicles and energy metabolism. Clin. Chim. Acta 2019, 488, 116–121.
- Rodríguez, M.; Silva, J.; Herrera, A.; Herrera, M.; Peña, C.; Martín, P.; Gil-Calderón, B.; Larriba, M.J.; Coronado, M.J.; Soldevilla, B.; et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget 2015, 6, 40575–40587.
- Borges, F.T.; Melo, S.A.; Özdemir, B.C.; Kato, N.; Revuelta, I.; Miller, C.A.; Gattone, V.H., 2nd; LeBleu, V.S.; Kalluri, R. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J. Am. Soc. Nephrol. 2013, 24, 385–392.
- Nakamura, Y.; Miyaki, S.; Ishitobi, H.; Matsuyama, S.; Nakasa, T.; Kamei, N.; Akimoto, T.; Higashi, Y.; Ochi, M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015, 589, 1257–1265.
- Gartz, M.; Strande, J.L. Examining the Paracrine Effects of Exosomes in Cardiovascular Disease and Repair. J. Am. Heart Assoc. 2018, 7, e007954.
- Pegtel, D.M.; Peferoen, L.; Amor, S. Extracellular vesicles as modulators of cell-to-cell communication in the healthy and diseased brain. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130516.
- Bang, C.; Thum, T. Exosomes: New players in cell-cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064.
- Samanta, S.; Rajasingh, S.; Drosos, N.; Zhou, Z.; Dawn, B.; Rajasingh, J. Exosomes: New molecular targets of diseases. Acta Pharmacol. Sin. 2018, 39, 501–513.
- Emmanouilidou, E.; Melachroinou, K.; Roumeliotis, T.; Garbis, S.D.; Ntzouni, M.; Margaritis, L.H.; Stefanis, L.; Vekrellis, K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J. Neurosci. 2010, 30, 6838–6851.
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177.
- Saá, P.; Yakovleva, O.; de Castro, J.; Vasilyeva, I.; De Paoli, S.H.; Simak, J.; Cervenakova, L. First demonstration of transmissible spongiform encephalopathy-associated prion protein (PrPTSE) in extracellular vesicles from plasma of mice infected with mouse-adapted variant Creutzfeldt-Jakob disease by in vitro amplification. J. Biol. Chem. 2014, 289, 29247–29260.
- Basso, M.; Pozzi, S.; Tortarolo, M.; Fiordaliso, F.; Bisighini, C.; Pasetto, L.; Spaltro, G.; Lidonnici, D.; Gensano, F.; Battaglia, E.; et al. Mutant copper-zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: Implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J. Biol. Chem. 2013, 288, 15699–15711.
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; Lucci, A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721.
- Psaila, B.; Kaplan, R.N.; Port, E.R.; Lyden, D. Priming the ‘soil’ for breast cancer metastasis: The pre-metastatic niche. Breast Dis. 2007, 26, 65–74.
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335.
- Silva, J.; Garcia, V.; Rodriguez, M.; Compte, M.; Cisneros, E.; Veguillas, P.; Garcia, J.M.; Dominguez, G.; Campos-Martin, Y.; Cuevas, J.; et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer 2012, 51, 409–418.
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624, Erratum in Nat. Cell Biol. 2008, 10, 752.
- Choi, D.; Montermini, L.; Kim, D.K.; Meehan, B.; Roth, F.P.; Rak, J. The Impact of Oncogenic EGFRvIII on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol. Cell Proteom. 2018, 17, 1948–1964.
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126 Pt 24, 5553–5565.
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980.
- Tamai, K.; Tanaka, N.; Nakano, T.; Kakazu, E.; Kondo, Y.; Inoue, J.; Shiina, M.; Fukushima, K.; Hoshino, T.; Sano, K.; et al. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun. 2010, 399, 384–390.
- Gross, J.C.; Chaudhary, V.; Bartscherer, K.; Boutros, M. Active Wnt proteins are secreted on exosomes. Nat. Cell Biol. 2012, 14, 1036–1045.
- Valcz, G.; Galamb, O.; Krenács, T.; Spisák, S.; Kalmár, A.; Patai, Á.V.; Wichmann, B.; Dede, K.; Tulassay, Z.; Molnár, B. Exosomes in colorectal carcinoma formation: ALIX under the magnifying glass. Mod. Pathol. 2016, 29, 928–938.
- Yang, J.; Zhang, Y.; Gao, X.; Yuan, Y.; Zhao, J.; Zhou, S.; Wang, H.; Wang, L.; Xu, G.; Li, X.; et al. Plasma-Derived Exosomal ALIX as a Novel Biomarker for Diagnosis and Classification of Pancreatic Cancer. Front. Oncol. 2021, 11, 628346.
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, e201904113.
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428.
- Monypenny, J.; Milewicz, H.; Flores-Borja, F.; Weitsman, G.; Cheung, A.; Chowdhury, R.; Burgoyne, T.; Arulappu, A.; Lawler, K.; Barber, P.R.; et al. ALIX Regulates Tumor-Mediated Immunosuppression by Controlling EGFR Activity and PD-L1 Presentation. Cell Rep. 2018, 24, 630–641.
- Yang, Y.; Hong, Q.; Shi, P.; Liu, Z.; Luo, J.; Shao, Z. Elevated expression of syntenin in breast cancer is correlated with lymph node metastasis and poor patient survival. Breast Cancer Res. 2013, 15, R50.
- Das, S.K.; Guo, C.; Pradhan, A.K.; Bhoopathi, P.; Talukdar, S.; Shen, X.N.; Emdad, L.; Subler, M.A.; Windle, J.J.; Sarkar, D.; et al. Knockout of MDA-9/Syntenin (SDCBP) expression in the microenvironment dampens tumor-supporting inflammation and inhibits melanoma metastasis. Oncotarget 2016, 7, 46848–46861.
- Kim, O.; Hwangbo, C.; Tran, P.T.; Lee, J.H. Syntenin-1-mediated small extracellular vesicles promotes cell growth, migration, and angiogenesis by increasing onco-miRNAs secretion in lung cancer cells. Cell Death Dis. 2022, 13, 122.
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 2021, 23, 631–641.
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685.
- Tkachenko, E.; Rhodes, J.M.; Simons, M. Syndecans: New kids on the signaling block. Circ. Res. 2005, 96, 488–500.
- David, G.; Zimmermann, P. Heparanase tailors syndecan for exosome production. Mol. Cell Oncol. 2015, 3, e1047556.
- Gallagher, J. Fell-Muir Lecture: Heparan sulphate and the art of cell regulation: A polymer chain conducts the protein orchestra. Int. J. Exp. Pathol. 2015, 96, 203–231.
- Hammond, E.; Khurana, A.; Shridhar, V.; Dredge, K. The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front. Oncol. 2014, 4, 195.
- Ilan, N.; Elkin, M.; Vlodavsky, I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int. J. Biochem. Cell Biol. 2006, 38, 2018–2039.
- Mytilinaiou, M.; Nikitovic, D.; Berdiaki, A.; Kostouras, A.; Papoutsidakis, A.; Tsatsakis, A.M.; Tzanakakis, G.N. Emerging roles of syndecan 2 in epithelial and mesenchymal cancer progression. IUBMB Life 2017, 69, 824–833.
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 2013, 288, 10093–10099.
- Fares, J.; Kashyap, R.; Zimmermann, P. Syntenin: Key player in cancer exosome biogenesis and uptake? Cell Adh. Migr. 2017, 11, 124–126.
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606.
- Batagov, A.O.; Kurochkin, I.V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol. Direct. 2013, 8, 12.
- Yang, B.; Feng, X.; Liu, H.; Tong, R.; Wu, J.; Li, C.; Yu, H.; Chen, Y.; Cheng, Q.; Chen, J.; et al. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene 2020, 39, 6529–6543.
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24.
- Arenaccio, C.; Federico, M. The Multifaceted Functions of Exosomes in Health and Disease: An Overview. Adv. Exp. Med. Biol. 2017, 998, 3–19.
- Kalluri, R.; LeBleu, V.S. Discovery of Double-Stranded Genomic DNA in Circulating Exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280.
- Krug, A.K.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.G.; Castellanos-Rizaldos, E.; et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 2018, 29, 700–706.
- Castellanos-Rizaldos, E.; Grimm, D.G.; Tadigotla, V.; Hurley, J.; Healy, J.; Neal, P.L.; Sher, M.; Venkatesan, R.; Karlovich, C.; Raponi, M.; et al. Exosome-Based Detection of EGFR T790M in Plasma from Non-Small Cell Lung Cancer Patients. Clin. Cancer Res. 2018, 24, 2944–2950.
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450.
- Kim, D.K.; Lee, J.; Kim, S.R.; Choi, D.S.; Yoon, Y.J.; Kim, J.H.; Go, G.; Nhung, D.; Hong, K.; Jang, S.C.; et al. EVpedia: A community web portal for extracellular vesicles research. Bioinformatics 2015, 31, 933–939.
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692.
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112.
- Liu, T.; Zhang, Q.; Zhang, J.; Li, C.; Miao, Y.R.; Lei, Q.; Li, Q.; Guo, A.Y. EVmiRNA: A database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019, 47, D89–D93.
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524.
- Ketting, R.F. microRNA Biogenesis and Function: An overview. Adv. Exp. Med. Biol. 2011, 700, 1–14.
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179.
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer—A brief overview. Adv. Biol. Regul. 2015, 57, 1–9, Erratum in Adv. Biol. Regul. 2015, 58, 53.
- Santos, R.M.; Moreno, C.; Zhang, W.C. Non-Coding RNAs in Lung Tumor Initiation and Progression. Int. J. Mol. Sci. 2020, 16, 2774.
- Soares, E.; Reis, J.; Rodrigues, M.; Ribeiro, C.F.; Pereira, F.C. Circulating Extracellular Vesicles: The Missing Link between Physical Exercise and Depression Management? Int. J. Mol. Sci. 2021, 22, 542.
- Cheng, G. Circulating miRNAs: Roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. 2015, 81, 75–93.
- Schwarzenbach, H. Clinical Relevance of Circulating, Cell-Free and Exosomal microRNAs in Plasma and Serum of Breast Cancer Patients. Oncol. Res. Treat. 2017, 40, 423–429.
- Wu, L.; Zhou, W.B.; Zhou, J.; Wei, Y.; Wang, H.M.; Liu, X.D.; Chen, X.C.; Wang, W.; Ye, L.; Yao, L.C.; et al. Circulating exosomal microRNAs as novel potential detection biomarkers in pancreatic cancer. Oncol. Lett. 2020, 20, 1432–1440.
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319.
- Yagi, Y.; Ohkubo, T.; Kawaji, H.; Machida, A.; Miyata, H.; Goda, S.; Roy, S.; Hayashizaki, Y.; Suzuki, H.; Yokota, T. Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neurosci. Lett. 2017, 636, 48–57.
- Siciliano, V.; Garzilli, I.; Fracassi, C.; Criscuolo, S.; Ventre, S.; di Bernardo, D. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat. Commun. 2013, 4, 2364.
- Shore, S.; Henderson, J.M.; Lebedev, A.; Salcedo, M.P.; Zon, G.; McCaffrey, A.P.; Paul, N.; Hogrefe, R.I. Small RNA Library Preparation Method for Next-Generation Sequencing Using Chemical Modifications to Prevent Adapter Dimer Formation. PLoS ONE 2016, 11, e0167009.
- Buschmann, D.; Kirchner, B.; Hermann, S.; Märte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles 2018, 7, 1481321, Erratum in J. Extracell. Vesicles 2019, 8, 1581487.
- Li, W.J.; Chen, H.; Tong, M.L.; Niu, J.J.; Zhu, X.Z.; Lin, L.R. Comparison of the yield and purity of plasma exosomes extracted by ultracentrifugation, precipitation, and membrane-based approaches. Open Chem. 2022, 20, 182–191.
- Olivares, D.; Perez-Hernandez, J.; Perez-Gil, D.; Chaves, F.J.; Redon, J.; Cortes, R. Optimization of small RNA library preparation protocol from human urinary exosomes. J. Transl. Med. 2020, 18, 132.
- Loudig, O.; Liu, C.; Rohan, T.; Ben-Dov, I.Z. Retrospective MicroRNA Sequencing: Complementary DNA Library Preparation Protocol Using Formalin-fixed Paraffin-embedded RNA Specimens. J. Vis. Exp. 2018, 135, 57471.
- Loudig, O.; Wang, T.; Ye, K.; Lin, J.; Wang, Y.; Ramnauth, A.; Liu, C.; Stark, A.; Chitale, D.; Greenlee, R.; et al. Evaluation and Adaptation of a Laboratory-Based cDNA Library Preparation Protocol for Retrospective Sequencing of Archived MicroRNAs from up to 35-Year-Old Clinical FFPE Specimens. Int. J. Mol. Sci. 2017, 18, 627.
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325.
- Precazzini, F.; Detassis, S.; Imperatori, A.S.; Denti, M.A.; Campomenosi, P. Measurements Methods for the Development of MicroRNA-Based Tests for Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1176.
- Ryu, K.J.; Lee, J.Y.; Choi, M.E.; Yoon, S.E.; Cho, J.; Ko, Y.H.; Shim, J.H.; Kim, W.S.; Park, C.; Kim, S.J. Serum-Derived Exosomal MicroRNA Profiles Can Predict Poor Survival Outcomes in Patients with Extranodal Natural Killer/T-Cell Lymphoma. Cancers 2020, 12, 3548.
- Sanchez Herrero, J.F.; Pluvinet, R.; Luna de Haro, A.; Sumoy, L. Paired-end small RNA sequencing reveals a possible overestimation in the isomiR sequence repertoire previously reported from conventional single read data analysis. BMC Bioinform. 2021, 22, 215.
- Zelli, V.; Compagnoni, C.; Capelli, R.; Corrente, A.; Cornice, J.; Vecchiotti, D.; Di Padova, M.; Zazzeroni, F.; Alesse, E.; Tessitore, A. Emerging Role of isomiRs in Cancer: State of the Art and Recent Advances. Genes 2021, 12, 1447.
- Giallombardo, M.; Chacártegui Borrás, J.; Castiglia, M.; Van Der Steen, N.; Mertens, I.; Pauwels, P.; Peeters, M.; Rolfo, C. Exosomal miRNA Analysis in Non-small Cell Lung Cancer (NSCLC) Patients’ Plasma Through qPCR: A Feasible Liquid Biopsy Tool. J. Vis. Exp. 2016, 111, 53900.
- Rodríguez, M.; Bajo-Santos, C.; Hessvik, N.P.; Lorenz, S.; Fromm, B.; Berge, V.; Sandvig, K.; Linē, A.; Llorente, A. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 2017, 16, 156.
- Tang, Y.; Zhao, Y.; Song, X.; Song, X.; Niu, L.; Xie, L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J. Clin. Lab. Anal. 2019, 33, e23004.
- Yoon, J.H.; Choi, B.J.; Nam, S.W.; Park, W.S. Gastric cancer exosomes contribute to the field cancerization of gastric epithelial cells surrounding gastric cancer. Gastric Cancer 2022, 25, 490–502.
- White, T.B.; McCoy, A.M.; Streva, V.A.; Fenrich, J.; Deininger, P.L. A droplet digital PCR detection method for rare L1 insertions in tumors. Mob. DNA 2014, 5, 30.
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610.
- Zhu, G.; Ye, X.; Dong, Z.; Lu, Y.C.; Sun, Y.; Liu, Y.; McCormack, R.; Gu, Y.; Liu, X. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J. Mol. Diagn. 2015, 17, 265–272.
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005.
- Malumbres, M.; Carnero, A. Cell cycle deregulation: A common motif in cancer. Prog. Cell Cycle Res. 2003, 5, 5–18.
- Su, L.; Zhang, J.; Zhang, X.; Zheng, L.; Zhu, Z. Identification of cell cycle as the critical pathway modulated by exosome-derived microRNAs in gallbladder carcinoma. Med. Oncol. 2021, 38, 141.
- Xia, H.; Li, M.; Chen, L.; Leng, W.; Yuan, D.; Pang, X.; Chen, L.; Li, R.; Tang, Q.; Bi, F. Suppression of RND3 activity by AES downregulation promotes cancer cell proliferation and invasion. Int. J. Mol. Med. 2013, 31, 1081–1086.
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674.
- Razavipour, S.F.; Harikumar, K.B.; Slingerland, J.M. p27 as a Transcriptional Regulator: New Roles in Development and Cancer. Cancer Res. 2020, 80, 3451–3458.
- Zhang, Z.; Xing, T.; Chen, Y.; Xiao, J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-β1 exposure. Biomed. Pharmacother. 2018, 106, 1135–1143.
- Yan, S.; Liu, G.; Jin, C.; Wang, Z.; Duan, Q.; Xu, J.; Xu, D. MicroRNA-6869-5p acts as a tumor suppressor via targeting TLR4/NF-κB signaling pathway in colorectal cancer. J. Cell Physiol. 2018, 233, 6660–6668.
- Yan, S.; Han, B.; Gao, S.; Wang, X.; Wang, Z.; Wang, F.; Zhang, J.; Xu, D.; Sun, B. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 2017, 8, 60149–60158.
- Wang, M.; Yu, F.; Ding, H.; Wang, Y.; Li, P.; Wang, K. Emerging Function and Clinical Values of Exosomal MicroRNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 791–804.
- Avrutsky, M.I.; Troy, C.M. Caspase-9: A Multimodal Therapeutic Target With Diverse Cellular Expression in Human Disease. Front. Pharmacol. 2021, 12, 701301.
- Jing, X.; Xie, M.; Ding, K.; Xu, T.; Fang, Y.; Ma, P.; Shu, Y. Exosome-transmitted miR-769-5p confers cisplatin resistance and progression in gastric cancer by targeting CASP9 and promoting the ubiquitination degradation of p53. Clin. Transl. Med. 2022, 12, e780.
- Wei, Y.; Li, M.; Cui, S.; Wang, D.; Zhang, C.Y.; Zen, K.; Li, L. Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes. Molecules 2016, 21, 777.
- Kwon, Y.; Kim, M.; Kim, Y.; Jung, H.S.; Jeoung, D. Exosomal MicroRNAs as Mediators of Cellular Interactions between Cancer Cells and Macrophages. Front. Immunol. 2020, 11, 1167.
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460.
- Owen, J.L.; Mohamadzadeh, M. Macrophages and chemokines as mediators of angiogenesis. Front. Physiol. 2013, 4, 159.
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016, 7, 43076–43087.
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017, 38, 522–528.
- Deregowska, A.; Wnuk, M. RAP1/TERF2IP-A Multifunctional Player in Cancer Development. Cancers 2021, 13, 5970.
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145.
- Martin-Gayo, E.; Yu, X.G. Role of Dendritic Cells in Natural Immune Control of HIV-1 Infection. Front. Immunol. 2019, 10, 1306.
- Veglia, F.; Gabrilovich, D.I. Dendritic cells in cancer: The role revisited. Curr. Opin. Immunol. 2017, 45, 43–51.
- Romano, R.; Picca, A.; Eusebi, L.H.U.; Marzetti, E.; Calvani, R.; Moro, L.; Bucci, C.; Guerra, F. Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression. Cells 2021, 10, 1361.
- Zhou, M.; Chen, J.; Zhou, L.; Chen, W.; Ding, G.; Cao, L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell Immunol. 2014, 292, 65–69.
- Lundholm, M.; Schröder, M.; Nagaeva, O.; Baranov, V.; Widmark, A.; Mincheva-Nilsson, L.; Wikström, P. Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: Mechanism of immune evasion. PLoS ONE 2014, 9, e108925.
- Childs, R.W.; Carlsten, M. Therapeutic approaches to enhance natural killer cell cytotoxicity against cancer: The force awakens. Nat. Rev. Drug Discov. 2015, 14, 487–498.
- Thompson, T.W.; Kim, A.B.; Li, P.J.; Wang, J.; Jackson, B.T.; Huang, K.T.H.; Zhang, L.; Raulet, D.H. Endothelial cells express NKG2D ligands and desensitize antitumor NK responses. Elife 2017, 6, e30881.
- Briand, J.; Garnier, D.; Nadaradjane, A.; Clément-Colmou, K.; Potiron, V.; Supiot, S.; Bougras-Cartron, G.; Frenel, J.S.; Heymann, D.; Vallette, F.M.; et al. Radiotherapy-induced overexpression of exosomal miRNA-378a-3p in cancer cells limits natural killer cells cytotoxicity. Epigenomics 2020, 12, 397–408.
- Duan, S.; Guo, W.; Xu, Z.; He, Y.; Liang, C.; Mo, Y.; Wang, Y.; Xiong, F.; Guo, C.; Li, Y.; et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol. Cancer 2019, 18, 29.
- Berchem, G.; Noman, M.Z.; Bosseler, M.; Paggetti, J.; Baconnais, S.; Le Cam, E.; Nanbakhsh, A.; Moussay, E.; Mami-Chouaib, F.; Janji, B.; et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. Oncoimmunology 2015, 5, e1062968.
- Alečković, M.; Kang, Y. Regulation of cancer metastasis by cell-free miRNAs. Biochim. Biophys. Acta 2015, 1855, 24–42.
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G.; et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445.
- Cai, Q.; Zhu, A.; Gong, L. Exosomes of glioma cells deliver miR-148a to promote proliferation and metastasis of glioblastoma via targeting CADM1. Bull. Cancer 2018, 105, 643–651.
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194.
- Felicetti, F.; De Feo, A.; Coscia, C.; Puglisi, R.; Pedini, F.; Pasquini, L.; Bellenghi, M.; Errico, M.C.; Pagani, E.; Carè, A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016, 14, 56.
- Sai, B.; Dai, Y.; Fan, S.; Wang, F.; Wang, L.; Li, Z.; Tang, J.; Wang, L.; Zhang, X.; Zheng, L.; et al. Cancer-educated mesenchymal stem cells promote the survival of cancer cells at primary and distant metastatic sites via the expansion of bone marrow-derived-PMN-MDSCs. Cell Death Dis. 2019, 10, 941.
- Zhang, X.; Sai, B.; Wang, F.; Wang, L.; Wang, Y.; Zheng, L.; Li, G.; Tang, J.; Xiang, J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer 2019, 18, 40.
- Théry, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular Characterization of Dendritic Cell-Derived Exosomes. Selective Accumulation of the Heat Shock Protein Hsc73. J. Cell Biol. 1999, 147, 599–610.
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct from Apoptotic Vesicles. J. Immunol. 2001, 166, 7309–7318.
- van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf-Bensussan, N.; Heyman, M. Intestinal Epithelial Cells Secrete Exosome-like Vesicles. Gastroenterology 2001, 121, 337–349.
- Skokos, D.; Botros, H.G.; Demeure, C.; Morin, J.; Peronet, R.; Birkenmeier, G.; Boudaly, S.; Mécheri, S. Mast Cell-Derived Exosomes Induce Phenotypic and Functional Maturation of Dendritic Cells and Elicit Specific Immune Responses In Vivo. J. Immunol. Baltim. Md. 2003, 170, 3037–3045.
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, E.; Pap, E.; Kittel, A.; et al. Membrane Vesicles, Current State-of-the-Art: Emerging Role of Extracellular Vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688.
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289.
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066.
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188.
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17.
- Teng, F.; Fussenegger, M. Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 2021, 8, 2003505.
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977.
- Marar, C.; Starich, B.; Wirtz, D. Extracellular Vesicles in Immunomodulation and Tumor Progression. Nat. Immunol. 2021, 22, 560–570.
- Cheng, L.; Hill, A.F. Therapeutically Harnessing Extracellular Vesicles. Nat. Rev. Drug Discov. 2022, 21, 379–399.
- Østergaard, O.; Nielsen, C.T.; Iversen, L.V.; Jacobsen, S.; Tanassi, J.T.; Heegaard, N.H.H. Quantitative Proteome Profiling of Normal Human Circulating Microparticles. J. Proteome Res. 2012, 11, 2154–2163.
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS ONE 2015, 10, e0145686.
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A Comparison of Methods for the Isolation and Separation of Extracellular Vesicles from Protein and Lipid Particles in Human Serum. Sci. Rep. 2020, 10, 1039.
- Askeland, A.; Borup, A.; Østergaard, O.; Olsen, J.V.; Lund, S.M.; Christiansen, G.; Kristensen, S.R.; Heegaard, N.H.H.; Pedersen, S. Mass-Spectrometry Based Proteome Comparison of Extracellular Vesicle Isolation Methods: Comparison of ME-Kit, Size-Exclusion Chromatography, and High-Speed Centrifugation. Biomedicines 2020, 8, 246.
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977.
- Kasprzyk, J.; Stępień, E.; Piekoszewski, W. Application of Nano-LC-MALDI-TOF/TOF-MS for Proteomic Analysis of Microvesicles. Clin. Biochem. 2017, 50, 241–243.
- Fel, A.; Lewandowska, A.E.; Petrides, P.E.; Wiśniewski, J.R. Comparison of Proteome Composition of Serum Enriched in Extracellular Vesicles Isolated from Polycythemia Vera Patients and Healthy Controls. Proteomes 2019, 7, 20.
- Chan, J.C.; Morgan, C.P.; Adrian Leu, N.; Shetty, A.; Cisse, Y.M.; Nugent, B.M.; Morrison, K.E.; Jašarević, E.; Huang, W.; Kanyuch, N.; et al. Reproductive Tract Extracellular Vesicles Are Sufficient to Transmit Intergenerational Stress and Program Neurodevelopment. Nat. Commun. 2020, 11, 1499.
- Zougman, A.; Selby, P.J.; Banks, R.E. Suspension Trapping (STrap) Sample Preparation Method for Bottom-up Proteomics Analysis. Proteomics 2014, 14, 1006–1000.
- Wu, C.; Zhou, S.; Mitchell, M.I.; Hou, C.; Byers, S.; Loudig, O.; Ma, J. Coupling Suspension Trapping–Based Sample Preparation and Data-Independent Acquisition Mass Spectrometry for Sensitive Exosomal Proteomic Analysis. Anal. Bioanal. Chem. 2022, 414, 2585–2595.
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic Insights and Diagnostic Potential. Expert Rev. Proteom. 2009, 6, 267–283.
- Schey, K.L.; Luther, J.M.; Rose, K.L. Proteomics Characterization of Exosome Cargo. Methods 2015, 87, 75–82.
- Kreimer, S.; Belov, A.M.; Ghiran, I.; Murthy, S.K.; Frank, D.A.; Ivanov, A.R. Mass-Spectrometry-Based Molecular Characterization of Extracellular Vesicles: Lipidomics and Proteomics. J. Proteome Res. 2015, 14, 2367–2384.
- Choi, D.-S.; Kim, D.-K.; Kim, Y.-K.; Gho, Y.S. Proteomics of Extracellular Vesicles: Exosomes and Ectosomes. Mass Spectrom. Rev. 2015, 34, 474–490.
- Rosa-Fernandes, L.; Rocha, V.B.; Carregari, V.C.; Urbani, A.; Palmisano, G. A Perspective on Extracellular Vesicles Proteomics. Front. Chem. 2017, 5, 102.
- Geis-Asteggiante, L.; Dhabaria, A.; Edwards, N.; Ostrand-Rosenberg, S.; Fenselau, C. Top–down Analysis of Low Mass Proteins in Exosomes Shed by Murine Myeloid-Derived Suppressor Cells. Int. J. Mass Spectrom. 2015, 378, 264–269.
- Fujita, K.; Kume, H.; Matsuzaki, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Uemura, M.; Miyagawa, Y.; Tomonaga, T.; Nonomura, N. Proteomic Analysis of Urinary Extracellular Vesicles from High Gleason Score Prostate Cancer. Sci. Rep. 2017, 7, 42961.
- Fricke, F.; Michalak, M.; Warnken, U.; Hausser, I.; Schnölzer, M.; Kopitz, J.; Gebert, J. SILAC-Based Quantification of TGFBR2-Regulated Protein Expression in Extracellular Vesicles of Microsatellite Unstable Colorectal Cancers. Int. J. Mol. Sci. 2019, 20, E4162.
- Kulkarni, S.; Koller, A.; Mani, K.M.; Wen, R.; Alfieri, A.; Saha, S.; Wang, J.; Patel, P.; Bandeira, N.; Guha, C.; et al. Identifying Urinary and Serum Exosome Biomarkers For Radiation Exposure Using A DDA and SWATH-MS Combined Workflow. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 566–577.
- Ole, N.; Jensen, O.N. Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 2006, 7, 391–403.
- Muller, M. Post-Translational Modifications of Protein Backbones: Unique Functions, Mechanisms, and Challenges. Biochemistry 2018, 57, 177–185.
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-Scale Proteomics and Phosphoproteomics of Urinary Exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379.
- Saraswat, M.; Joenväära, S.; Musante, L.; Peltoniemi, H.; Holthofer, H.; Renkonen, R. N-Linked (N-) Glycoproteomics of Urinary Exosomes. Mol. Cell. Proteom. 2015, 14, 2298.
- Brown, C.J.; Gaunitz, S.; Wang, Z.; Strindelius, L.; Jacobson, S.C.; Clemmer, D.E.; Trinidad, J.C.; Novotny, M.V. Glycoproteomic Analysis of Human Urinary Exosomes. Anal. Chem. 2020, 92, 14357–14365.
- Chen, I.-H.; Xue, L.; Hsu, C.-C.; Paez, J.S.P.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.-K.; Tao, W.A. Phosphoproteins in Extracellular Vesicles as Candidate Markers for Breast Cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180.
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic Profiling of Extracellular Vesicles Allows for Human Breast Cancer Subtyping. Commun. Biol. 2019, 2, 325.
- Lo Cicero, A.; Stahl, P.D.; Raposo, G. Extracellular Vesicles Shuffling Intercellular Messages: For Good or for Bad. Curr. Opin. Cell Biol. 2015, 35, 69–77.
- Yu, R.K.; Tsai, Y.-T.; Ariga, T. Functional Roles of Gangliosides in Neurodevelopment: An Overview of Recent Advances. Neurochem. Res. 2012, 37, 1230–1244.
- Sunshine, H.; Iruela-Arispe, M.L. Membrane lipids and cell signaling. Curr. Opin. Lipidol. 2017, 28, 408–413.
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293.
- Gould, S.B. Membranes and evolution. Curr. Biol. 2018, 28, R381–R385.
- Singer, S.J.; Nicolson, G.L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731.
- Jackson Catherine, L.; Walch, L.; Verbavatz, J.-M. Lipids and Their Trafficking: An Integral Part of Cellular Organization. Dev. Cell 2016, 39, 139–153.
- Kang, J.X.; Wan, J.B.; He, C. Concise Review: Regulation of Stem Cell Proliferation and Differentiation by Essential Fatty Acids and Their Metabolites. Stem Cells 2014, 32, 1092–1098.
- Wang, Q.; Song, Y.H.; Tang, Z.; Wang, Z.P.; Xu, Q.; Bao, N. Effects of ganglioside GM1 and neural growth factor on neural stem cell proliferation and differentiation. Genet. Mol. Res. 2016, 15, 1–10.
- Newton, J.; Lima, S.; Maceyka, M.; Spiegel, S. Revisiting the sphingolipid rheostat: Evolving concepts in cancer therapy. Exp. Cell Res. 2015, 333, 195–200.
- Houten, S.M.; Wanders, R.J.A. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 2010, 33, 469–477.
- Abuhusain, H.J.; Matin, A.; Qiao, Q.; Shen, H.; Kain, N.; Day, B.W.; Stringer, B.W.; Daniels, B.; Laaksonen, M.A.; Teo, C.; et al. A metabolic shift favoring sphingosine 1-phosphate at the expense of ceramide controls glioblastoma angiogenesis. J. Biol. Chem. 2013, 288, 37355–37364.
- Koizumi, A.; Narita, S.; Nakanishi, H.; Ishikawa, M.; Eguchi, S.; Kimura, H.; Takasuga, S.; Huang, M.; Inoue, T.; Sasaki, J.; et al. Increased fatty acyl saturation of phosphatidylinositol phosphates in prostate cancer progression. Sci. Rep. 2019, 9, 13257.
- Greenlee, J.D.; Subramanian, T.; Liu, K.; King, M.R. Rafting Down the Metastatic Cascade: The Role of Lipid Rafts in Cancer Metastasis, Cell Death, and Clinical Outcomes. Cancer Res. 2021, 81, 5.
- Sriram, V.; Cho, S.; Li, P.; O’Donnell Patrick, W.; Dunn, C.; Hayakawa, K.; Blum Janice, S.; Brutkiewicz Randy, R. Inhibition of glycolipid shedding rescues recognition of a CD1+ T cell lymphoma by natural killer T (NKT) cells. Proc. Natl. Acad. Sci. USA 2002, 99, 8197–8202.
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18.
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41.
- Wang, W.; Zhu, N.; Yan, T.; Shi, Y.-N.; Chen, J.; Zhang, C.-J.; Xie, X.-J.; Liao, D.-F.; Qin, L. The crosstalk: Exosomes and lipid metabolism. Cell Commun. Signal. 2020, 18, 119.
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220.
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321.
- Zebrowska, A.; Skowronek, A.; Wojakowska, A.; Widlak, P.; Pietrowska, M. Metabolome of Exosomes: Focus on Vesicles Released by Cancer Cells and Present in Human Body Fluids. Int. J. Mol. Sci. 2019, 20, 3461.
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309.
- Yi, X.; Li, Y.; Hu, X.; Wang, F.; Liu, T. Changes in phospholipid metabolism in exosomes of hormone-sensitive and hormone-resistant prostate cancer cells. J. Cancer 2021, 12, 2893–2902.
- Lydic, T.A.; Townsend, S.; Adda, C.G.; Collins, C.; Mathivanan, S.; Reid, G.E. Rapid and comprehensive ‘shotgun’ lipidome profiling of colorectal cancer cell derived exosomes. Methods 2015, 87, 83–95.
- Haraszti, R.A.; Didiot, M.-C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570.
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Biochim. Biophys. Acta 2011, 1811, 637–647.
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.H.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.O.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14.
- Lobasso, S.; Tanzarella, P.; Mannavola, F.; Tucci, M.; Silvestris, F.; Felici, C.; Ingrosso, C.; Corcelli, A.; Lopalco, P. A Lipidomic Approach to Identify Potential Biomarkers in Exosomes from Melanoma Cells with Different Metastatic Potential. Front. Physiol. 2021, 12, 748895.
- Peterka, O.; Jirásko, R.; Chocholoušková, M.; Kuchař, L.; Wolrab, D.; Hájek, R.; Vrána, D.; Strouhal, O.; Melichar, B.; Holčapek, M. Lipidomic characterization of exosomes isolated from human plasma using various mass spectrometry techniques. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158634.
- Altadill, T.; Campoy, I.; Lanau, L.; Gill, K.; Rigau, M.; Gil-Moreno, A.; Reventos, J.; Byers, S.; Colas, E.; Cheema, A.K. Enabling Metabolomics Based Biomarker Discovery Studies Using Molecular Phenotyping of Exosome-Like Vesicles. PLoS ONE 2016, 11, e0151339.
- Xu, S.; Ye, M.; Xu, D.; Li, X.; Pan, C.; Zou, H. Matrix with high salt tolerance for the analysis of peptide and protein samples by desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2006, 78, 2593–2599.
- Tran, A.; Wan, L.; Xu, Z.; Haro, J.M.; Li, B.; Jones, J.W. Lithium Hydroxide Hydrolysis Combined with MALDI TOF Mass Spectrometry for Rapid Sphingolipid Detection. J. Am. Soc. Mass. Spectrom. 2021, 32, 289–300.
- Paglia, G.; Kliman, M.; Claude, E.; Geromanos, S.; Astarita, G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal. Bioanal. Chem. 2015, 407, 4995–5007.
- Bowman, A.P.; Blakney, G.T.; Hendrickson, C.L.; Ellis, S.R.; Heeren, R.M.A.; Smith, D.F. Ultra-High Mass Resolving Power, Mass Accuracy, and Dynamic Range MALDI Mass Spectrometry Imaging by 21-T FT-ICR MS. Anal. Chem. 2020, 92, 3133–3142.
- Dodds, J.N.; Baker, E.S. Ion Mobility Spectrometry: Fundamental Concepts, Instrumentation, Applications, and the Road Ahead. J. Am. Soc. Mass Spectrom. 2019, 30, 2185–2195.
- Giles, K.; Ujma, J.; Wildgoose, J.; Pringle, S.; Richardson, K.; Langridge, D.; Green, M. A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem. 2019, 91, 8564–8573.
- Claude, E.; Jones, E.A.; Pringle, S.D. DESI Mass Spectrometry Imaging (MSI). Methods Mol. Biol. 2017, 1618, 65–75.
- Han, X.; Gross, R.W. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass. Spectrom. Rev. 2005, 24, 367–412.
- Hsu, F.-F. Mass spectrometry-based shotgun lipidomics—A critical review from the technical point of view. Anal. Bioanal. Chem. 2018, 410, 6387–6409.
- Su, B.; Bettcher, L.F.; Hsieh, W.-Y.; Hornburg, D.; Pearson, M.J.; Blomberg, N.; Giera, M.; Snyder, M.P.; Raftery, D.; Bensinger, S.J.; et al. A DMS Shotgun Lipidomics Workflow Application to Facilitate High-Throughput, Comprehensive Lipidomics. J. Am. Soc. Mass Spectrom. 2021, 32, 2655–2663.
- Wojakowska, A.; Zebrowska, A.; Skowronek, A.; Rutkowski, T.; Polanski, K.; Widlak, P.; Marczak, L.; Pietrowska, M. Metabolic Profiles of Whole Serum and Serum-Derived Exosomes Are Different in Head and Neck Cancer Patients Treated by Radiotherapy. J. Pers. Med. 2020, 10, 229.
- Fuchs, B.; Süß, R.; Teuber, K.; Eibisch, M.; Schiller, J. Lipid analysis by thin-layer chromatography—A review of the current state. J. Chromatogr. A 2011, 1218, 2754–2774.
- Engel, K.M.; Prabutzki, P.; Leopold, J.; Nimptsch, A.; Lemmnitzer, K.; Vos, D.R.N.; Hopf, C.; Schiller, J. A new update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 2022, 86, 101145.
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Anal. Chem. TRAC 2014, 61, 192–206.
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic Analysis. Anal. Chem. 2018, 90, 4249–4257.
- Lange, M.; Fedorova, M. Evaluation of lipid quantification accuracy using HILIC and RPLC MS on the example of NIST® SRM® 1950 metabolites in human plasma. Anal. Bioanal. Chem. 2020, 412, 3573–3584.
- Rampler, E.; Criscuolo, A.; Zeller, M.; El Abiead, Y.; Schoeny, H.; Hermann, G.; Sokol, E.; Cook, K.; Peake, D.A.; Delanghe, B.; et al. A Novel Lipidomics Workflow for Improved Human Plasma Identification and Quantification Using RPLC-MSn Methods and Isotope Dilution Strategies. Anal. Chem. 2018, 90, 6494–6501.
- Jung, J.H.; Lee, M.Y.; Choi, D.Y.; Lee, J.W.; You, S.; Lee, K.Y.; Kim, J.; Kim, K.P. Phospholipids of tumor extracellular vesicles stratify gefitinib-resistant nonsmall cell lung cancer cells from gefitinib-sensitive cells. Proteomics 2015, 15, 824–835.
- Aiello, N.M.; Brabletz, T.; Kang, Y.; Nieto, M.A.; Weinberg, R.A.; Stanger, B.Z. Upholding a role for EMT in pancreatic cancer metastasis. Nature 2017, 547, E7–E8.
- Zhou, P.; Li, B.; Liu, F.; Zhang, M.; Wang, Q.; Liu, Y.; Yao, Y.; Li, D. The epithelial to mesenchymal transition (EMT) and cancer stem cells: Implication for treatment resistance in pancreatic cancer. Mol. Cancer 2017, 16, 52.
- Brzozowski, J.S.; Jankowski, H.; Bond, D.R.; McCague, S.B.; Munro, B.R.; Predebon, M.J.; Scarlett, C.J.; Skelding, K.A.; Weidenhofer, J. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018, 17, 211.
- Cheng, L.; Zhang, K.; Qing, Y.; Li, D.; Cui, M.; Jin, P.; Xu, T. Proteomic and lipidomic analysis of exosomes derived from ovarian cancer cells and ovarian surface epithelial cells. J. Ovarian Res. 2020, 13, 9.
- Kuc, N.; Doermann, A.; Shirey, C.; Lee, D.D.; Lowe, C.-W.; Awasthi, N.; Schwarz, R.E.; Stahelin, R.V.; Schwarz, M.A. Pancreatic ductal adenocarcinoma cell secreted extracellular vesicles containing ceramide-1-phosphate promote pancreatic cancer stem cell motility. Biochem. Pharmacol. 2018, 156, 458–466.
- Fan, T.W.M.; Zhang, X.; Wang, C.; Yang, Y.; Kang, W.-Y.; Arnold, S.; Higashi, R.M.; Liu, J.; Lane, A.N. Exosomal lipids for classifying early and late stage non-small cell lung cancer. Anal. Chim. Acta 2018, 1037, 256–264.
- Tao, L.; Zhou, J.; Yuan, C.; Zhang, L.; Li, D.; Si, D.; Xiu, D.; Zhong, L. Metabolomics identifies serum and exosomes metabolite markers of pancreatic cancer. Metabolomics 2019, 15, 86.
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132.
- Kang, Y.-T.; Purcell, E.; Palacios-Rolston, C.; Lo, T.-W.; Ramnath, N.; Jolly, S.; Nagrath, S. Isolation and Profiling of Circulating Tumor-Associated Exosomes Using Extracellular Vesicular Lipid-Protein Binding Affinity Based Microfluidic Device. Small 2019, 15, e1903600.
- Matsumura, S.; Minamisawa, T.; Suga, K.; Kishita, H.; Akagi, T.; Ichiki, T.; Ichikawa, Y.; Shiba, K. Subtypes of tumour cell-derived small extracellular vesicles having differently externalized phosphatidylserine. J. Extracell. Vesicles 2019, 8, 1579541.
- Sharma, R.; Huang, X.; Brekken, R.A.; Schroit, A.J. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br. J. Cancer 2017, 117, 545–552.
- Nakai, W.; Yoshida, T.; Diez, D.; Miyatake, Y.; Nishibu, T.; Imawaka, N.; Naruse, K.; Sadamura, Y.; Hanayama, R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016, 6, 33935.
- Shih, C.-L.; Chong, K.-Y.; Hsu, S.-C.; Chien, H.-J.; Ma, C.-T.; Chang, J.W.-C.; Yu, C.-J.; Chiou, C.-C. Development of a magnetic bead-based method for the collection of circulating extracellular vesicles. New Biotechnol. 2016, 33, 116–122.
- Vallabhapurapu, S.D.; Blanco, V.M.; Sulaiman, M.K.; Vallabhapurapu, S.L.; Chu, Z.; Franco, R.S.; Qi, X. Variation in human cancer cell external phosphatidylserine is regulated by flippase activity and intracellular calcium. Oncotarget 2015, 6, 34375–34388.
- Utsugi, T.; Schroit, A.J.; Connor, J.; Bucana, C.D.; Fidler, I.J. Elevated Expression of Phosphatidylserine in the Outer Membrane Leaflet of Human Tumor Cells and Recognition by Activated Human Blood Monocytes. Cancer Res. 1991, 51, 3062–3066.
- Riedl, S.; Rinner, B.; Asslaber, M.; Schaider, H.; Walzer, S.; Novak, A.; Lohner, K.; Zweytick, D. In search of a novel target—Phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochim. Biophys. Acta 2011, 1808, 2638–2645.
- Dong, H.P.; Holth, A.; Kleinberg, L.; Ruud, M.G.; Elstrand, M.B.; Tropé, C.G.; Davidson, B.; Risberg, B. Evaluation of cell surface expression of phosphatidylserine in ovarian carcinoma effusions using the annexin-V/7-AAD assay: Clinical relevance and comparison with other apoptosis parameters. Am. J. Clin. Pathol. 2009, 132, 756–762.
- Kelleher, R.J., Jr.; Balu-Iyer, S.; Loyall, J.; Sacca, A.J.; Shenoy, G.N.; Peng, P.; Iyer, V.; Fathallah, A.M.; Berenson, C.S.; Wallace, P.K.; et al. Extracellular Vesicles Present in Human Ovarian Tumor Microenvironments Induce a Phosphatidylserine-Dependent Arrest in the T-cell Signaling Cascade. Cancer Immunol. Res. 2015, 3, 1269–1278.
- Lea, J.; Sharma, R.; Yang, F.; Zhu, H.; Ward, E.S.; Schroit, A.J. Detection of phosphatidylserine-positive exosomes as a diagnostic marker for ovarian malignancies: A proof of concept study. Oncotarget 2017, 8, 14395–14407.
This entry is offline, you can click here to edit this entry!
