Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Deubiquitinating enzymes (DUBs) are a group of proteases that are important for maintaining cell homeostasis by regulating the balance between ubiquitination and deubiquitination. As the only known metalloproteinase family of DUBs, JAB1/MPN/Mov34 metalloenzymes (JAMMs) are specifically associated with tumorigenesis and immunological and inflammatory diseases at multiple levels. The far smaller numbers and distinct catalytic mechanism of JAMMs render them attractive drug targets. Several JAMM inhibitors have been successfully developed and have shown promising therapeutic efficacy.
- deubiquitinating enzymes
- JAMMs
- structural basis
- catalytic mechanism
- functions
- inhibitors
1. Introduction
Protein ubiquitination, defined as a process that covalently conjugates ubiquitin to the target protein, is one of the most powerful post-translational modifications regulating virtually all cellular processes, such as cell death, cell cycle, and DNA repair [1][2][3][4]. Ubiquitin is a 76-amino-acid, 8-kDa polypeptide with a conserved sequence that is present universally and ubiquitously in eukaryotes [5][6]. Full-length ubiquitin contains eight ubiquitination sites, including seven lysine residues (K6, K11, K27, K29, K33, K48, K63) and an N-terminal methionine residue (M1) (Figure 1A) [7][8]. Under the sequential action of ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3), an isopeptide linkage is formed between the carboxyl group of the ubiquitin C-terminal glycine and the ε-amino group of the target protein lysine (Figure 1B) [9][10]. Then, the Gly 76 of additional ubiquitin molecules (called distal ubiquitin) can be covalently attached to the ubiquitination sites in ubiquitin itself (called proximal ubiquitin), to produce polyubiquitin chains [11]. As a result, various types of protein ubiquitination are formed, which determine the fate of ubiquitinated substrates [6][12]. For example, polyubiquitin chains linked via the K48 of internal ubiquitin groups are used for protein degradation signaling by the ubiquitin-proteasome system (Figure 1C1,C2), whereas K63-linked polyubiquitin chains, presenting different architecture, play proteasome-independent roles in various intracellular events, such as inflammatory signaling, DNA repair, ribosomal protein synthesis, endocytosis, and vesicular trafficking (Figure 1D1,D2) [13][14][15]. Additionally, some other ubiquitin-like modifications, such as small ubiquitin-like modifier (SUMO), neuronal precursor cell-expressed developmentally downregulated protein 8 (NEDD8), and interferon stimulated gene 15 (ISG15), can also be ligated to target proteins in a process similar to ubiquitylation, mostly to provide nondegradative signals [16].
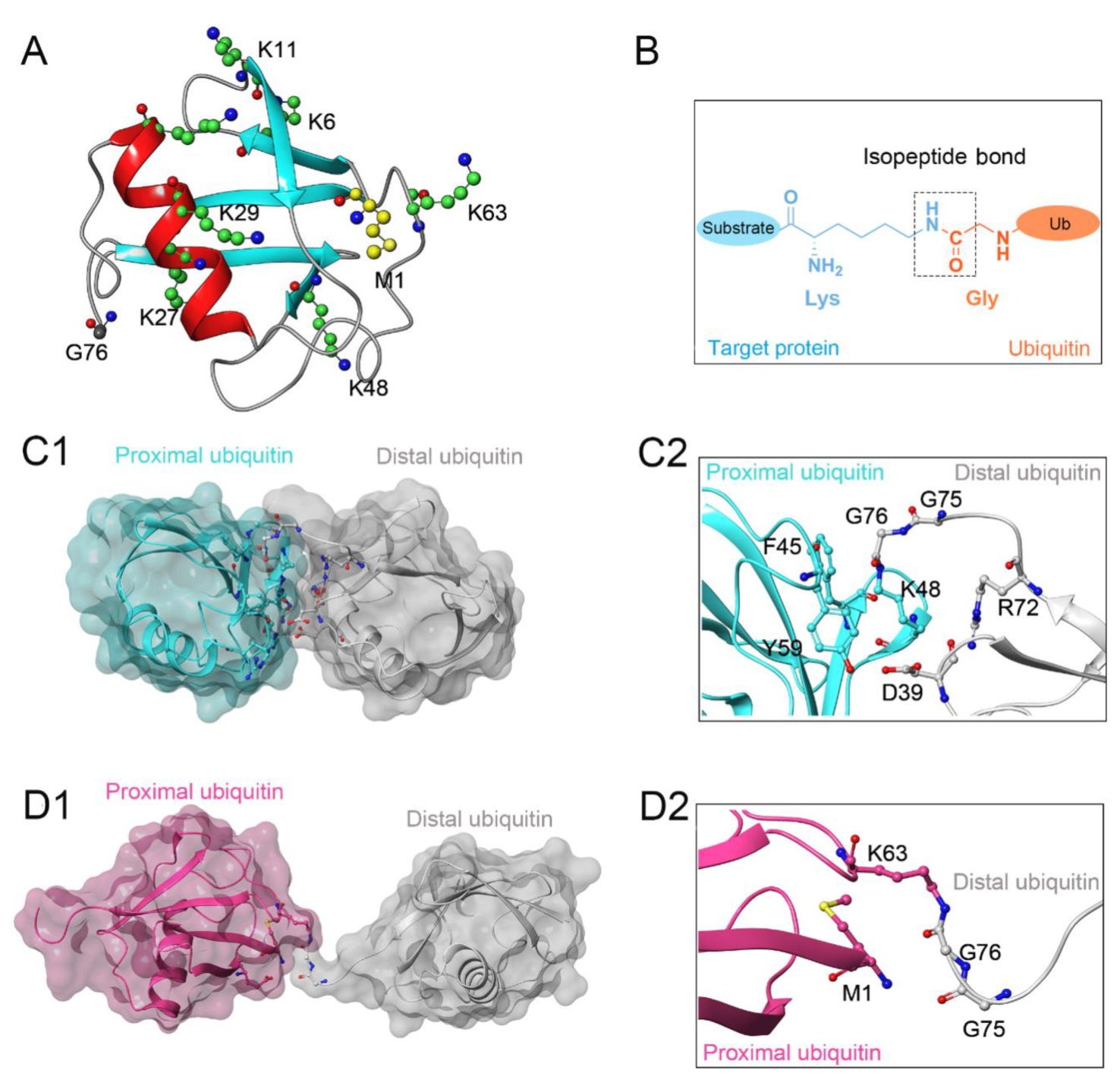
Figure 1. (A) Crystal structure of human ubiquitin (PDB ID: 1UBQ). Seven lysine residues and an N-terminal methionine residue are colored green and yellow, respectively. (B) The isopeptide bond between the ubiquitin glycine residue (orange) and the target protein lysine residue (blue). (C1,C2) The overall structure and local conformation of K48-linked polyubiquitin chains (PDB ID: 1TBE). (D1,D2) The overall structure and local conformation of K63-linked polyubiquitin chains (PDB ID: 3HM3). The distal and proximal ubiquitin are colored gray and cyan, respectively.
To date, the human genome encodes nearly 600 E3 ligases while only approximately 100 deubiquitinating enzymes (DUBs) have been identified, clustered in the following 7 families: 56 ubiquitin-specific peptidases (USPs), 17 ovarian tumor proteases (OTUs), 12 JAB1/MPN/Mov34 metalloenzymes (JAMMs), 5 motif interacting with ubiquitin-containing novel DUB family proteases (MINDYs), 4 ubiquitin C-terminal hydroxylases (UCHs), 4 Machado-Josephin domain proteases (MJDs), and 1 zinc finger-containing ubiquitin peptidase 1 (ZUP1) [17][18][19]. Six of these seven families are cysteine proteases, whereas only the JAMM family are zinc-dependent metalloproteinases. In the case of cysteine protease DUBs, the catalytic domains contain a highly conserved catalytic triad comprising cysteine and nearby histidine and aspartate residues [8][20]. In contrast, metalloprotease DUBs coordinate Zn2+ ion with histidine, aspartate, and serine residues to attack the isopeptide bond by activating a water molecule [21].
Dysfunction of the ubiquitin system, especially DUBs, has been recognized as a contributing factor in the origin of many human diseases, such as cancer, inflammatory diseases, and neurological diseases [22][23]. Notably, there has been a recent expansion of drug discovery programs targeting JAMMs. Unlike the large number of cysteine protease DUBs (~90), as few as 12 JAMMs have been identified in the human genome, among which only 7 (AMSH, AMSH-LP, BRCC36, eIF3h, Rpn11, MYSM1, and CSN5) exhibit isopeptidase activity toward ubiquitin chains [17][24]. Furthermore, multiple JAMM-related signaling pathways, such as DNA damage control (BRCC36) [25], endocytosis (AMSH, AMSH-LP) [26][27], protein biosynthesis (eIF3h) [28], and protein degradation (Rpn11, CSN5) [29], have been confirmed to be associated with numerous diseases, including tumorigenesis and immunological and inflammatory disorders. The much fewer numbers, distinct catalytic mechanism, and, specifically, association with diseases render JAMMs a new class of potential drug targets [30]. To gain an in-depth understanding of JAMMs, this entry emphatically discusses the structural basis, catalytic mechanism, and diverse functions with a focus on JAMM family proteins, including AMSH, AMSH-LP, BRCC36, Rpn11, and CSN5. Researchers also summarize the current reported inhibitors targeting JAMM family members.
2. Structural Characteristic of JAMMs
The MPN (Mpr1/Pad1 N-terminal) domain is a striking characteristic of the JAMM family. In 2004, Ambroggio et al. first reported the crystal structure of the MPN domain protein (PDB ID: 1R5X) from a prokaryotic organism Archaeoglobus fulgidus AF2198 (AfJAMM) [31]. They revealed that the MPN domain of AfJAMM consisted of an eight-stranded β sheet (β1–β8), flanked by a long α helix (α1) between the first and second strand, and a short α helix (α2) between the fourth and fifth strand (Figure 2A). Subsequently, an increasing number of crystal structures were resolved and the MPN domain proteins could then be further divided into two subfamilies: (1) the MPN+ family, with isopeptidase activity, characterized by a zinc-coordinating JAMM motif (ExnHxHx7Sx2D) (where x represents any amino acid residue) (Figure 2K); and (2) the MPN– family, without catalytic activity, serving as scaffolds in some JAMM multi-subunit complexes [32][33][34].
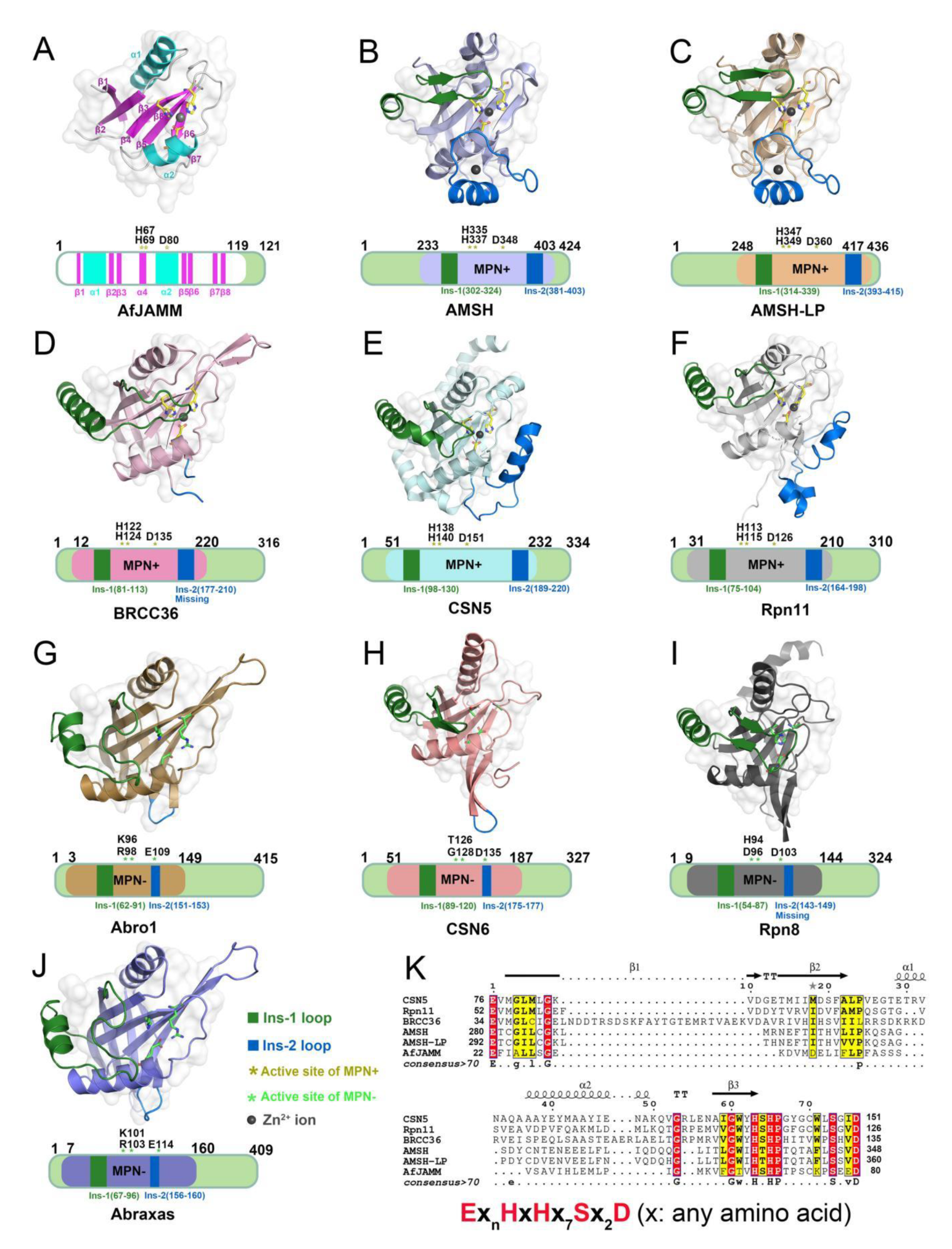
Figure 2. Structural characteristics of JAMM MPN domain mentioned in this entry. (A) Crystal structure of AfJAMM (PDB ID: 1R5X). AfJAMM has a typical MPN domain containing an eight-stranded β sheet (β1–β8) (fuchsia), a long α helix (α1), and a short α helix (α2) (cyan). (B–J) Crystal structure of AMSH (PDB ID: 3RZU) (light blue), AMSH-LP (PDB ID: 2ZNV) (wheat), BRCC36 (PDB ID: 6H3C) (light pink), CSN5 (PDB ID: 4F7O) (pale cyan), Rpn11 (PDB ID: 4O8X) (gray), Abro1 (PDB ID: 6H3C) (sand), CSN6 (PDB ID: 4D10) (salmon), Rpn8 (PDB ID: 4O8X) (light black), and Abraxas (PDB ID: 6GVW) (slate) MPN domain. All the Ins-1 and Ins-2 loop are colored deep green and blue, respectively. The yellow and green asterisks represent active sites of MPN+ and MPN–, respectively. The black round represents Zn2+ ion. (K) The zinc-coordinating JAMM motif of MPN+ (ExnHxHx7Sx2D) (where x is any amino acid residue).
Most JAMMs possess two unique insertions, referred to as Ins-1 and Ins-2, which are considered to play important roles in the recognition and binding of ubiquitinated protein substrates [24]. The Ins-1 segment forms one ridge of the substrate-binding groove to assist in the proper positioning of the C-terminal ubiquitin tail for catalysis while the Ins-2 region contributes to the productive substrate positioning [35].
3. Catalytic Mechanism of JAMMs
So far, 7 of the 12 JAMMs (AMSH, AMSH-LP, BRCC36, eIF3h, Rpn11, CSN5, and MYSM1) in the human genome belong to the MPN+ subfamily and have DUB activity toward proteins while the remaining 5 JAMMs (Abraxas, Abro1, CSN6, eIF3f, and Rpn8) belong to the MPN− subfamily [36][37]. Interestingly, most of these JAMMs require the formation of multi-subunit complexes to exert their isopeptidase activities, including Rpn11 and Rpn8 of the 26S proteasome [29], CSN5 and CSN6 of the COP9 signalosome [38], eIF3f and eIF3h of the human translation initiation factor 3 (eIF3) [39], BRCC36 and Abraxas of the BRCA1-A complex [40], and BRCC36 and Abro1 of the BRISC complex [41]. There are, of course, exceptions, such as AMSH and AMSH-LP, which can cleave K63-linked ubiquitin chains independent of protein partners [42]. Sato et al. resolved the crystal structure of AMSH-LPE292A-ubiquitin complex (PDB ID: 2ZNV) from H. sapiens and proposed the catalytic mechanism of JAMMs, which was probably similar to that of thermolysin [26][43].
First, the zinc-bound catalytic water is deprotonated by an active site Glu 292 and subsequently performs a nucleophilic attack on the substrate peptide carbonyl. Then, the negative charge on the peptide carbonyl oxygen is stabilized by the Zn2+ ion and His 347, His 349, Ser 357, and Asp 360 while the positive charge on the amide nitrogen is stabilized by Glu 292. The reaction then proceeds through an oxyanion tetrahedral intermediate and a second transition state, which results in the cleavage of the peptide N-C bond. With the proton transferring from the amide nitrogen to water, the cleavage of the peptide bond is ultimately completed [26]. Although the whole amino acid sequences of these seven MPN+ members are highly divergent, the catalytic core region is completely conserved, suggesting that they might employ identical catalytic mechanisms [30].
4. Structural and Functional Basis of JAMMs
4.1. Functional Basis of AMSH in Receptor Endocytosis
It has recently been shown that AMSH plays a significant role in regulating the endosomal sorting of many cell-surface receptors, which is a highly regulated process for maintaining cellular homeostasis and generating adaptive responses to external stimuli [44][45]. Typically, the endocytic trafficking process involves the internalization, endosomal sorting, and lysosomal degradation of cell-surface receptors and is strictly executed by the endosomal sorting complexes required for transport (ESCRT), consisting of at least five macromolecular assemblies termed ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III and vacuolar sorting protein 4 (Vps4) [46][47][48]. It is during this process that AMSH can interact with the components ESCRT-0 and ESCRT-III and so affect the fate of receptors [16].
Several studies have documented the crucial role of AMSH-mediated deubiquitination in the trafficking of endocytosed receptors, such as receptor-tyrosine kinase epidermal growth factor receptor (EGFR), G protein-coupled receptors (GPCRs), connexins 43 (connexin Cx43), and the inflammasome component NACHT, LRR, and PYD domain-containing protein (NALP7) (Table 1) [49][50][51][52][53][54]. For example, the E3-ligase c-Cbl has been shown to promote lysosomal degradation of the K63 ubiquitylated EGFR [55] while AMSH opposes this action and promotes EGFR recycling, thus regulating the balance of the intracellular EGFR content [51]. In another study, Ribeiro-Rodrigues et al. demonstrated that AMSH could protect gap junctions from degradation by mediating the deubiquitination of Cx43 to regulate intercellular communication [52]. By linking the DUBs to immune regulation, Mallampalli et al. found that AMSH cleaved K63-linked ubiquitin from NALP7 to increase its intracellular content, leading to inflammasome-dependent IL-1β cleavage and release [54]. For some important GPCRs, including chemokine receptor CXCR4, protease-activated receptor 2 (PAR2), and δ-opioid receptor (DOR), AMSH has been reported to regulate their stability and trafficking, as the loss of AMSH catalytic activity can significantly alter the steady-state level of GPCRs [49][50][53]. Overall, AMSH-mediated receptor endocytosis is accomplished through the recognition of specific ubiquitination patterns, specifically multi-monoubiquitination and K63-linked polyubiquitination.
Table 1. List of JAMMs mentioned in this entry and their functional roles.
| Proteins | Functional Complex | Target Protein | Linkage Type | Regulation Effects | Ref. |
|---|---|---|---|---|---|
| AMSH | N/A | EGFR | K63- | Promote the recycling of EGFR | [51] |
| AMSH | N/A | Cx43 | K63- | Protect gap junctions from degradation to regulate the intercellular communication | [52] |
| AMSH | N/A | NALP7 | K63- | Lead to the inflammasome-dependent IL-1β cleavage and release | [54] |
| AMSH | N/A | CXCR4 | Mono- | Regulate the stability and trafficking of CXCR4 | [53] |
| AMSH | N/A | PAR2 | Mono- | Regulate the trafficking and down-regulation of PAR2 | [49] |
| AMSH | N/A | DOR | Mono- | Regulate the downregulation of the DOR | [50] |
| BRCC36 | BRISC | NLRP3 | K63- | Activate NLRP3 and promote inflammasome assembly | [56] |
| BRCC36 | BRISC | IFNAR1/2 | K63- | Promote the cellular response to Type I interferons | [57] |
| BRCC36 | BRISC | HIV-1 Tat | K63- | Rescue Tat from destruction to potentiate the effectiveness of antiviral regimens | [58] |
| BRCC36 | BRISC | NuMA | K63- | Promote the assembly of functional bipolar spindle during mitosis | [59] |
| BRCC36 | BRISC | JAK2 | K63- | Limit hematopoietic stem cell expansion | [60] |
| BRCC36 | BRCA1-A | H2A/H2AX | K63- | Suppress hyperactive HR repair | [61] |
| Rpn11 | 26S proteasome | c-Jun | K48- | Maintain a stable intracellular concentration of c-Jun | [62] |
| Rpn11 | 26S proteasome | E2F1 | K63- | Stabilize E2F1 protein to promote tumorigenesis | [63] |
| Rpn11 | 26S proteasome | ErbB2 | ND | Regulate ErbB2 ubiquitylation and stability in cancer cells | [64] |
| Rpn11 | 26S proteasome | H2A/H2AX | K63- | Promote the correct coordination of the cellular response to DSB | [65] |
| Rpn11 | 26S proteasome | Mitf | ND | Allow more stable Mitf expression in osteoclast differentiation process | [66] |
| CSN5 | Cop9 signalosome | CRLs | NEDD8 | Maintain the proper activity of CRLs in myriad cellular processes | [67] |
N/A: Not applicable; ND: Not determined.
4.2. Structural Basis of AMSH
ESCRT-0 mainly comprises two subunits, Hrs and STAM [68]. The interaction of ESCRT-0 and AMSH is achieved through the binding of the SH3 domain of STAM with the SH3-binding motif (SBM) of AMSH [69]. Based on the NMR structure of AMSH and STAM complex (PDB ID: 5IXF) from H. sapiens, Hologne et al. considered that the interaction of AMSH-SBM and STAM-SH3 contributed to the correct positioning of polyubiquitin chains toward AMSH before cleavage [70][71]. Subsequently, they proposed a structural model for AMSH-STAM-ubiquitin complex, in which the activation of AMSH was allowed by facile, simultaneous binding to two ubiquitin groups in a polyubiquitin substrate: one (distal ubiquitin) by the catalytic domain of the AMSH and the other (proximal ubiquitin) by the UIM domain of STAM. Such a binding mode would stabilize the ubiquitin chain in a productive orientation, resulting in an enhancement of the DUB activity [70][71]. Meanwhile, another ubiquitin-binding domain of STAM, the VHS domain, is shown to enhance the cleavage of ubiquitin chains composed of more than two ubiquitin molecules. The absence of the VHS domain removes the specificity toward tri-ubiquitin, suggesting that this domain is essential for specificity toward longer chains [71].
The assembly of ESCRT-III is a highly ordered process involving seven charged multivesicular body protein (CHMP) subunits (CHMP 1-7) [72]. The N-terminus of AMSH contains nuclear localization signal and the microtubule-interacting and transport (MIT) domain, which has been shown to interact with several CHMPs, including CHMP1, CHMP2, and CHMP3 [45][73]. However, in the case of the crystal structure of an N-terminal fragment of AMSH (AMSHΔC) in complex with the C-terminal region of CHMP3 (CHMP3ΔN) (PDB ID: 2XZE) from H. sapiens, Solomons et al. found a higher affinity between CHMP3 and AMSH, indicating that AMSH might employ different interaction surfaces for these CHMPs [74]. They also found that CHMP3ΔN interacted with the MIT domain of AMSH involving multiple amino acids, including a hydrogen bond between Glu 207 and Tyr 80 and salt bridges between Glu 203-Lys 88, Arg 216-Glu 104, and Arg 221-Glu 72. The CHMP3 C-terminal residue Ser 222 was capped by AMSHΔC Lys 107, contributing to the formation of a salt bridge with the carboxyl group of Ser 222 and hydrogen bonds with the carbonyls of Thr 219 and Leu 220. Given this, an appropriate molecular model of the AMSH-CHMP3 complex is proposed, in which AMSH is first recruited to membranes early in the ESCRT pathway via ESCRT-0 STAM [75]. Then, the helical extension of the AMSH MIT domain can serve as a long arm to position the DUB activity > 20 nm away from the ESCRT-III polymer, thus reaching into the vesicle formed by ESCRT-I and ESCRT-II for receptor deubiquitination [26][74].
4.3. Comparison of AMSH and AMSH-LP
Interestingly, AMSH has a close homolog AMSH-LP (AMSH-like protein) [35]. Although the entire amino acid sequences of AMSH and AMSH-LP are only 54% identical, their three-dimensional structures exhibit extremely high similarity [16][26]. Especially, their catalytic domains and residues involved in proximal ubiquitin recognition are completely conserved [42][76]. However, AMSH-LP lacks several key features and presents some significant differences in the residues used for interaction with the distal ubiquitin [77]. Besides, AMSH contains an SBM domain, which interacts with the STAM of ESCRT-0 while a functional SBM is lost in human AMSH-LP [16]. By further exploring the differences in the properties, Davies et al. found that the catalytic domain of AMSH was thermodynamically less stable than that of AMSH-LP. They suggested that a more stable protein (AMSH-LP) was likely to have improved close packing of side chains, making it more rigid, whereas a less stable protein (AMSH) would be more plastic, which may make AMSH more suitable for interacting with other proteins, such as ESCRT-0 and ESCRT-III [77].
5. Currently Reported Inhibitors Targeting JAMMs
Recently, increasingly more attention has been placed on JAMMs and many inhibitors have been designed to target AMSH (BC-1471) [54], Rpn11 (8TQ, capzimin, thiolutin, holomycin, SOP6, SOP11, O-phenanthroline) [78][79][80][81], and CSN5 (Berberine, CSN5i-3) (Figure 3A) [82][83].
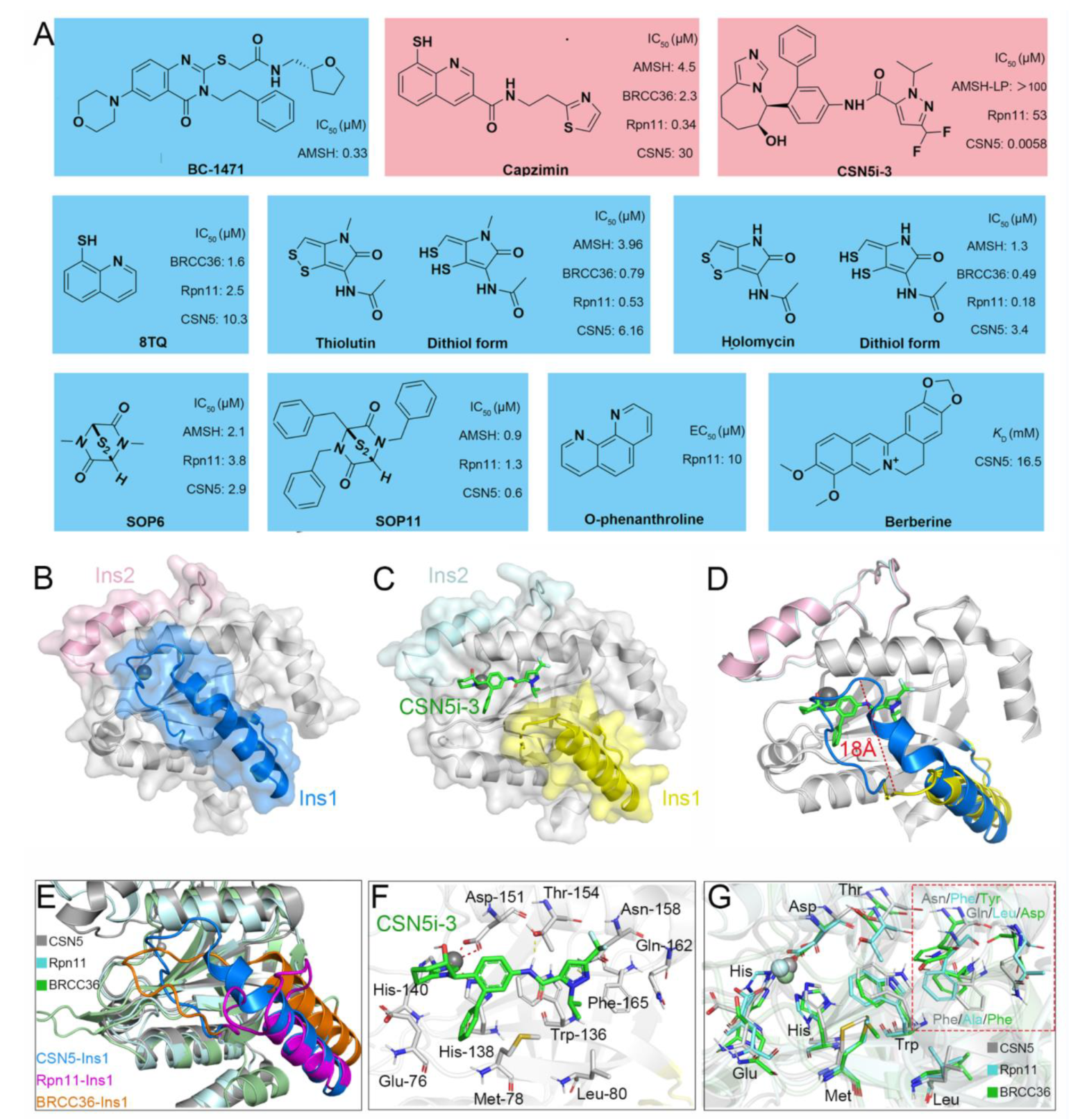
Figure 3. (A) Previously reported JAMM inhibitors. The Rpn11-specific inhibitor capzimin and CSN5-specific inhibitor CSN5i-3 are colored red. (B) Crystal structure of CSN5 (PDB ID: 4D10). The Ins-1 and Ins-2 are colored blue and pink, respectively. (C,F) Crystal structure of CSN5 in complex with CSN5i-3 (PDB ID: 5JOG). The Ins-1 and Ins-2 are colored yellow and palecyan, respectively. The CSN5i-3 is colored light green. (D) Superposition of the crystal structures of CSN5 and the complex of CSN5 and CSN5i-3. (E,G) Superposition of the crystal structures of CSN5, Rpn11, and BRCC36 (PDB ID: 6H3C). The CSN5, Rpn11, and BRCC36 are colored gray, cyan, and green, respectively. The Ins-1 regions of CSN5, Rpn11, and BRCC36 are colored blue, purple, and orange, respectively.
AMSH. Typically, after ubiquitination of the NALP7 inflammasome at Lys 288 and/or Lys 290, it is rapidly recruited by STAM for the subsequent endosomal passage and lysosomal degradation [84][85]. However, under the deubiquitination of AMSH, NALP7 is rescued from endolysosome sorting to permit inflammasome-dependent IL-1β cleavage [16]. Given this, by targeting the AMSH-ubiquitin-binding pocket, Bednash et al. performed a computer-assisted virtual screening using a library containing more than 500,000 experimental compounds [54]. As a result, a small molecule BC-1471 (IC50 = 0.33 μM) was observed to selectively decrease NALP7 abundance to suppress IL-1β release in several complementary human inflammatory systems, including THP-1 monocyte/macrophages, peripheral blood mononuclear cells, and lung organ culture. By evaluating BC-1471 against 38 other individual DUBs, mainly from cysteine families, the compound showed excellent specificity for AMSH because of no off-target DUB inhibition at the tested concentration. Afterwards, based on the molecular docking model of AMSH and BC-1471, the detailed mechanism of this interaction was further explored: Thr 63 formed electrostatic interactions with BC-1471, Tyr 105 formed π–π interactions with the benzene ring, and Val 97 formed σ–π interactions with the quinazoline ring. Together, these results indicate that BC-1471 exerts DUB inhibitory activity on AMSH to inhibit NALP7 inflammasome activity.
Rpn11. Multiple myeloma (MM) is a heterogeneous plasma cell malignancy for which there is currently no cure while the inhibition of the proteasome emerges as a powerful strategy for MM therapy [86][87]. Currently, the FDA has approved three medications, including bortezomib, carfilzomib, and ixazomib, to inhibit the proteasome by binding preferentially to the catalytic threonine residue of the β5 subunit within the 20S CP [88][89][90][91]. However, a proportion of patients do not respond to these compounds and those who do tend to relapse [92]. Therefore, there is an urgent need to develop new drugs targeting proteostasis with different mechanisms. Unlike the canonical proteasome inhibition, Rpn11-mediated inhibition occurs at 19S RP and thus may provide alternative opportunities to treat MM [29]. Importantly, it has been shown that Rpn11 is more highly expressed in patient MM cells while its loss-of-function by siRNA knockdown decreases MM cell viability [80].
In 2017, Li et al. and Perez et al. first identified a potent and selective moiety 8-thioquinoline (8TQ) (IC50 = 2.5 μM) by screening a library of metal-binding pharmacophores, which displayed strong inhibition of Rpn11 [81][93]. In addition, they demonstrated that 8TQ exerted its inhibitory activity through chelating the metal coordination of the active site Zn2+ ion. Unfortunately, 8TQ did not distinguish Rpn11 from other JAMMs, such as BRCC36 (IC50 = 1.6 μM) and CSN5 (IC50 = 10.3 μM). Subsequently, they performed structural optimization of 8TQ to further improve its inhibitory activity and selectivity. As a result, a lead compound capzimin was successfully developed, which showed potent activity for Rpn11 (IC50 = 0.34 µM) and high selectivity over other JAMMs such as AMSH (IC50 = 4.5 μM), BRCC36 (IC50 = 2.3 μM), and CSN5 (IC50 = 30 μM). Encouragingly, capzimin was equipotent against a set of bortezomib-sensitive and -resistant retinal pigment epithelial cells. Furthermore, capzimin also inhibited proliferation and induced apoptosis in several kinds of cancer cells, such as leukemia cells (SR and K562) and solid tumors (NCI-4460 and MCF7).
Initially, thiolutin (THL) was described as a dithiolopyrrolone antibiotic secreted by Streptomyces, and subsequently Lauinger et al. characterized it as a Zn2+ ion chelator capable of inhibiting DUB activity of several JAMMs, including AMSH (IC50 = 3.96 μM), BRCC36 (IC50 = 0.79 μM), Rpn11 (IC50 = 0.53 μM), and CSN5 (IC50 = 6.16 μM) [94]. Interestingly, THL exerted its activity only when it was reduced to the dithiol form inside the cell, which was similar to other dithiolopyrrolone antibiotics [95]. Moreover, the natural methyl derivative of THL, termed holomycin (HOL), was observed to inhibit Rpn11 (IC50 = 0.18 μM) and BRCC36 (IC50 = 0.49 μM) even more efficiently. Therefore, to a certain extent, THL and HOL might be regarded as selective inhibitors of BRCC36 and Rpn11. Afterwards, THL was used in the inhibition of BRCC36-mediated NLRP3 deubiquitination and activation by the Yin group [78]. They demonstrated that THL alleviated NLRP3-related inflammatory diseases in multiple mouse models, containing lipopolysaccharide-induced sepsis, monosodium urate-induced peritonitis, experimental autoimmune encephalomyelitis, CAPS, and methionine-choline-deficient diet-induced nonalcoholic fatty liver disease. As anticipated, HOL also displayed an even higher inhibitory activity against NLRP3 than THL. Molecular docking suggested that THL and HOL associated directly with Zn2+ ion of the BRCC36 active site, thus displacing the zinc-bound water molecule that was critical for the catalytic process. Additionally, the lower docking score of HOL (−6.392 kcal/mol) than THL (−5.169 kcal/mol) explained the reason for the higher inhibitory activity of HOL for BRCC36.
In 2018, Li et al. reported another type of Rpn11 inhibitor epidithiodiketopiperazines (ETPs), which were usually served as virulence factors generated from Aspergillus fumigatus secondary metabolites [79]. Among all tested ETPs, SOP6 was considered the core scaffold compound but did not show obvious selectivity between different JAMMs, such as AMSH (IC50 = 2.1 μM), Rpn11 (IC50 = 3.8 μM), and CSN5 (IC50 = 2.9 μM). Another ETP compound, SOP11, presented slightly higher inhibitory activity than SOP6 in AMSH (IC50 = 0.9 μM), Rpn11 (IC50 = 1.3 μM), and CSN5 (IC50 = 0.6 μM). Similar to capzimin, SOP11 not only triggered an unfolded protein response and induced an accumulation of polyubiquitin conjugates but also did not inhibit zinc-dependent enzymes unrelated to Rpn11, such as human carbonic anhydrase and matrix metalloproteinase 2 [79][81][93]. Meanwhile, the inhibition of proteasome by SOP11 had no effects on CSN5 activity, thus conferring it as a promising starting point to develop Rpn11 inhibitors. In another case of a known zinc chelator, O-phenanthroline (OPA) (EC50 = 10 μM) was also shown to induce apoptosis in MM cells and overcome resistance to the inhibitor bortezomib, which was ascribed to selectively inhibit Rpn11 activity without affecting other DUBs (e.g., USP1/USP2/USP4/USP5/USP7/USP8/USP20/UCH37) [80].
CSN5. CSN5 has been found to be overexpressed in a variety of cancers, including breast, thyroid, skin, ovarian, lung, and liver cancers [96], while the knockdown of CSN5 by siRNA has been shown to inhibit cell cycle progression and cause strong induction of apoptosis in hepatocellular carcinoma cells [97]. Because of this, stabilization of the neddylated CRLs through inhibition of CSN5 represents a novel therapeutic approach for the treatment of CSN5-dependent cancers. Based on the time-resolved fluorescence resonance energy transfer assay, researchers at Novartis developed a high-throughput screening platform for targeted screening of CSN5 inhibitors [82][98]. The final compound, CSN5i-3, inhibited CSN-mediated CRL deneddylation with an extremely low IC50 value (0.0058 μM) and showed a good pharmacokinetic profile. More importantly, CSN5i-3 had excellent selectivity since other JAMMs such as AMSH-LP (IC50 > 100 μM) and RPN11 (IC50 = 53 μM) were not or only weakly inhibited. In a panel of 500 cancer cell lines, CSN5i-3 exhibited varying degrees of inhibitory activities, and suppressed the growth of a human xenograft in mice.
Berberine (BBR), an isoquinoline quaternary alkaloid extracted from Coptis chinensis, has been used as a therapeutic agent in multiple diseases and presents strong anti-proliferative effects on cancer cells, such as breast, liver, and colorectal cancer cells [99][100][101]. By surface plasmon resonance assay, Liu et al. revealed a previously unrecognized antitumor mechanism of BBR: it could interact with CSN5 directly (KD = 16.25 mM) to inhibit its deneddylase activity, therefore triggering the proteasome-dependent degradation of PD-L1 and activating the tumor-infiltrating T cells [83]. The reduction in PD-L1 expression in the tumor microenvironment subsequently attenuated the activation of immunosuppressive myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs), resulting in the antitumor effect in Lewis tumor xenograft mice. Collectively, BBR is a small-molecule inhibitor of CSN5 that disrupted PD-L1-mediated immunosuppression.
This entry is adapted from the peer-reviewed paper 10.3390/biom12070910
References
- Imai, Y.; Soda, M.; Takahashi, R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 2000, 275, 35661–35664.
- Karin, M.; Ben-Neriah, Y. Phosphorylation meets ubiquitination: The control of NF-B activity. Annu. Rev. Immunol. 2000, 18, 621–663.
- Pagano, M.; Tam, S.W.; Theodoras, A.M.; Beer-Romero, P.; Del Sal, G.; Chau, V.; Yew, P.R.; Draetta, G.F.; Rolfe, M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 1995, 269, 682–685.
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428.
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880.
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229.
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586.
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563.
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422.
- Hershko, A.; Ciechanover, A.; Varshavsky, A. Basic medical research award. The ubiquitin system. Nat. Med. 2000, 6, 1073–1081.
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533.
- Kwon, Y.T.; Ciechanover, A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 2017, 42, 873–886.
- Hospenthal, M.K.; Freund, S.M.; Komander, D. Assembly, analysis and architecture of atypical ubiquitin chains. Nat. Struct. Mol. Biol. 2013, 20, 555–565.
- Ye, Y.; Blaser, G.; Horrocks, M.H.; Ruedas-Rama, M.J.; Ibrahim, S.; Zhukov, A.A.; Orte, A.; Klenerman, D.; Jackson, S.E.; Komander, D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 2012, 492, 266–270.
- Erpapazoglou, Z.; Walker, O.; Haguenauer-Tsapis, R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells 2014, 3, 1027–1088.
- Clague, M.J.; Urbé, S. Endocytosis: The DUB version. Trends Cell Biol. 2006, 16, 551–559.
- Schauer, N.J.; Magin, R.S.; Liu, X.; Doherty, L.M.; Buhrlage, S.J. Advances in discovering deubiquitinating enzyme (DUB) inhibitors. J. Med. Chem. 2020, 63, 2731–2750.
- Kwasna, D.; Abdul Rehman, S.A.; Natarajan, J.; Matthews, S.; Madden, R.; De Cesare, V.; Weidlich, S.; Virdee, S.; Ahel, I.; Gibbs-Seymour, I.; et al. Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol. Cell 2018, 70, 150–164.e156.
- Mevissen, T.E.T.; Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017, 86, 159–192.
- Hanpude, P.; Bhattacharya, S.; Dey, A.K.; Maiti, T.K. Deubiquitinating enzymes in cellular signaling and disease regulation. IUBMB Life 2015, 67, 544–555.
- Sowa, M.E.; Bennett, E.J.; Gygi, S.P.; Harper, J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009, 138, 389–403.
- Gadhave, K.; Kumar, P.; Kapuganti, S.K.; Uversky, V.N.; Giri, R. Unstructured biology of proteins from ubiquitin-proteasome system: Roles in cancer and neurodegenerative diseases. Biomolecules 2020, 10, 796.
- Cockram, P.E.; Kist, M.; Prakash, S.; Chen, S.H.; Wertz, I.E.; Vucic, D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021, 28, 591–605.
- Ronau, J.A.; Beckmann, J.F.; Hochstrasser, M. Substrate specificity of the ubiquitin and Ubl proteases. Cell Res. 2016, 26, 441–456.
- Patterson-Fortin, J.; Shao, G.; Bretscher, H.; Messick, T.E.; Greenberg, R.A. Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J. Biol. Chem. 2010, 285, 30971–30981.
- Sato, Y.; Yoshikawa, A.; Yamagata, A.; Mimura, H.; Yamashita, M.; Ookata, K.; Nureki, O.; Iwai, K.; Komada, M.; Fukai, S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 2008, 455, 358–362.
- McCullough, J.; Clague, M.J.; Urbé, S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cel. Biol. 2004, 166, 487–492.
- Marchione, R.; Leibovitch, S.A.; Lenormand, J.L. The translational factor eIF3f: The ambivalent eIF3 subunit. Cell Mol. Life Sci. 2013, 70, 3603–3616.
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R.; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615.
- Birol, M.; Echalier, A. Structure and function of MPN (Mpr1/PadN-terminal) domain-containing proteins. Curr. Protein Pept. Sci. 2014, 15, 504–517.
- Ambroggio, X.I.; Rees, D.C.; Deshaies, R.J. JAMM: A metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004, 2, 0113–0119.
- Dambacher, C.M.; Worden, E.J.; Herzik, M.A.; Martin, A.; Lander, G.C. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife 2016, 5, e13027.
- Galej, W.P.; Nguyen, T.H.D.; Newman, A.J.; Nagai, K. Structural studies of the spliceosome: Zooming into the heart of the machine. Curr. Opin. Struct. Biol. 2014, 25, 57–66.
- Lingaraju, G.M.; Bunker, R.D.; Cavadini, S.; Hess, D.; Hassiepen, U.; Renatus, M.; Fischer, E.S.; Thomä, N.H. Crystal structure of the human COP9 signalosome. Nature 2014, 512, 161–165.
- Shrestha, R.K.; Ronau, J.A.; Davies, C.W.; Guenette, R.G.; Strieter, E.R.; Paul, L.N.; Das, C. Insights into the mechanism of deubiquitination by JAMM deubiquitinases from cocrystal structures of the enzyme with the substrate and product. Biochemistry 2014, 53, 3199–3217.
- Fraile, J.M.; Quesada, V.; Rodríguez, D.; Freije, J.M.P.; López-Otín, C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene 2012, 31, 2373–2388.
- Maytal-Kivity, V.; Reis, N.; Hofmann, K.; Glickman, M.H. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002, 3, 28.
- Liu, Y.; Shah, S.V.; Xiang, X.; Wang, J.; Deng, Z.-B.; Liu, C.; Zhang, L.; Wu, J.; Edmonds, T.; Jambor, C.; et al. COP9-associated CSN5 regulates exosomal protein deubiquitination and sorting. Am. J. Pathol. 2009, 174, 1415–1425.
- Iadevaia, V.; Caldarola, S.; Tino, E.; Amaldi, F.; Loreni, F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA 2008, 14, 1730–1736.
- Dong, Y.; Hakimi, M.-A.; Chen, X.; Kumaraswamy, E.; Cooch, N.S.; Godwin, A.K.; Shiekhattar, R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell 2003, 12, 1087–1099.
- Zeqiraj, E.; Tian, L.; Piggott, C.A.; Pillon, M.C.; Duffy, N.M.; Ceccarelli, D.F.; Keszei, A.F.A.; Lorenzen, K.; Kurinov, I.; Orlicky, S.; et al. Higher-order assembly of BRCC36-KIAA0157 is required for DUB activity and biological function. Mol. Cell 2015, 59, 970–983.
- Nakamura, M.; Tanaka, N.; Kitamura, N.; Komada, M. Clathrin anchors deubiquitinating enzymes, AMSH and AMSH-like protein, on early endosomes. Genes Cells 2006, 11, 593–606.
- Holden, H.M.; Tronrud, D.E.; Monzingo, A.F.; Weaver, L.H.; Matthews, B.W. Slow- and fast-binding inhibitors of thermolysin display different modes of binding: Crystallographic analysis of extended phosphonamidate transition-state analogues. Biochemistry 1987, 26, 8542–8553.
- Wright, M.H.; Berlin, I.; Nash, P.D. Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination. Cell Biochem. Biophys. 2011, 60, 39–46.
- Kyuuma, M.; Kikuchi, K.; Kojima, K.; Sugawara, Y.; Sato, M.; Mano, N.; Goto, J.; Takeshita, T.; Yamamoto, A.; Sugamura, K.; et al. AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct. Funct. 2007, 31, 159–172.
- Agromayor, M.; Martin-Serrano, J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 2006, 281, 23083–23091.
- Shields, S.B.; Piper, R.C. How ubiquitin functions with ESCRTs. Traffic 2011, 12, 1306–1317.
- Henne, W.M.; Stenmark, H.; Emr, S.D. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 2013, 5, 1288–1302.
- Hasdemir, B.; Murphy, J.E.; Cottrell, G.S.; Bunnett, N.W. Endosomal deubiquitinating enzymes control ubiquitination and down-regulation of protease-activated receptor. J. Biol. Chem. 2009, 284, 28453–28466.
- Hislop, J.N.; Henry, A.G.; Marchese, A.; von Zastrow, M. Ubiquitination regulates proteolytic processing of G protein-coupled receptors after their sorting to lysosomes. J. Biol. Chem. 2009, 284, 19361–19370.
- Meijer, I.M.J.; van Rotterdam, W.; van Zoelen, E.J.J.; van Leeuwen, J.E.M. Recycling of EGFR and ErbB2 is associated with impaired Hrs tyrosine phosphorylation and decreased deubiquitination by AMSH. Cell. Signal. 2012, 24, 1981–1988.
- Ribeiro-Rodrigues, T.M.; Catarino, S.; Marques, C.; Ferreira, J.V.; Martins-Marques, T.; Pereira, P.; Girão, H. AMSH-mediated deubiquitination of Cx43 regulates internalization and degradation of gap junctions. FASEB J. 2014, 28, 4629–4641.
- Sierra, M.I.; Wright, M.H.; Nash, P.D. AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR. J. Biol. Chem. 2010, 285, 13990–14004.
- Bednash, J.S.; Weathington, N.; Londino, J.; Rojas, M.; Gulick, D.L.; Fort, R.; Han, S.; McKelvey, A.C.; Chen, B.B.; Mallampalli, R.K. Targeting the deubiquitinase STAMBP inhibits NALP7 inflammasome activity. Nat. Commun. 2017, 8, 15203.
- Levkowitz, G.; Waterman, H.; Zamir, E.; Kam, Z.; Oved, S.; Langdon, W.Y.; Beguinot, L.; Geiger, B.; Yarden, Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998, 12, 3663–3674.
- Py, B.F.; Kim, M.-S.; Vakifahmetoglu-Norberg, H.; Yuan, J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 2013, 49, 331–338.
- Zheng, H.; Gupta, V.; Patterson-Fortin, J.; Bhattacharya, S.; Katlinski, K.; Wu, J.; Varghese, B.; Carbone, C.J.; Aressy, B.; Fuchs, S.Y.; et al. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 2013, 5, 180–193.
- Xu, M.; Moresco, J.J.; Chang, M.; Mukim, A.; Smith, D.; Diedrich, J.K.; Yates, J.R.; Jones, K.A. SHMT2 and the BRCC36/BRISC deubiquitinase regulate HIV-1Tat K63-ubiquitylation and destruction by autophagy. PLoS Pathog. 2018, 14, e1007071.
- Yan, K.; Li, L.; Wang, X.; Hong, R.; Zhang, Y.; Yang, H.; Lin, M.; Zhang, S.; He, Q.; Zheng, D.; et al. The deubiquitinating enzyme complex BRISC is required for proper mitotic spindle assembly in mammalian cells. J. Cell Biol. 2015, 210, 209–224.
- Donaghy, R.; Han, X.; Rozenova, K.; Lv, K.; Jiang, Q.; Doepner, M.; Greenberg, R.A.; Tong, W. The BRISC deubiquitinating enzyme complex limits hematopoietic stem cell expansion by regulating JAKK63-ubiquitination. Blood 2019, 133, 1560–1571.
- Shao, G.; Lilli, D.R.; Patterson-Fortin, J.; Coleman, K.A.; Morrissey, D.E.; Greenberg, R.A. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl. Acad. Sci. USA 2009, 106, 3166–3171.
- Nabhan, J.F.; Ribeiro, P. The 19S proteasomal subunit POH1 contributes to the regulation of c-Jun ubiquitination, stability, and subcellular localization. J. Biol. Chem. 2006, 281, 16099–16107.
- Wang, B.; Ma, A.; Zhang, L.; Jin, W.-L.; Qian, Y.; Xu, G.; Qiu, B.; Yang, Z.; Liu, Y.; Xia, Q.; et al. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat. Commun. 2015, 6, 8704.
- Liu, H.; Buus, R.; Clague, M.J.; Urbé, S. Regulation of ErbB2 receptor status by the proteasomal DUB POH1. PLoS ONE 2009, 4, e5544.
- Butler, L.R.; Densham, R.M.; Jia, J.; Garvin, A.J.; Stone, H.R.; Shah, V.; Weekes, D.; Festy, F.; Beesley, J.; Morris, J.R. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 2012, 31, 3918–3934.
- Schwarz, T.; Sohn, C.; Kaiser, B.; Jensen, E.D.; Mansky, K.C. The 19S proteasomal lid subunit POH1 enhances the transcriptional activation by Mitf in osteoclasts. J. Cell. Biochem. 2010, 109, 967–974.
- Cope, G.A.; Deshaies, R.J. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 2003, 114, 663–671.
- Polo, S. Signaling-mediated control of ubiquitin ligases in endocytosis. BMC. Biol. 2012, 10, 25.
- McCullough, J.; Row, P.E.; Lorenzo, O.; Doherty, M.; Beynon, R.; Clague, M.J.; Urbé, S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 2006, 16, 160–165.
- Davies, C.W.; Paul, L.N.; Das, C. Mechanism of recruitment and activation of the endosome-associated deubiquitinase AMSH. Biochemistry 2013, 52, 7818–7829.
- Hologne, M.; Cantrelle, F.-X.; Riviere, G.; Guillière, F.; Trivelli, X.; Walker, O. NMR reveals the interplay among the AMSH SH3 binding motif, STAM2, and Lys63-linked diubiquitin. J. Mol. Biol. 2016, 428, 4544–4558.
- Azmi, I.F.; Davies, B.A.; Xiao, J.; Babst, M.; Xu, Z.; Katzmann, D.J. ESCRT-III family members stimulate Vps4ATPase activity directly or via Vta1. Dev. Cell 2008, 14, 50–61.
- Tsang, H.T.H.; Connell, J.W.; Brown, S.E.; Thompson, A.; Reid, E.; Sanderson, C.M. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics 2006, 88, 333–346.
- Solomons, J.; Sabin, C.; Poudevigne, E.; Usami, Y.; Hulsik, D.L.; Macheboeuf, P.; Hartlieb, B.; Göttlinger, H.; Weissenhorn, W. Structural basis for ESCRT-III CHMP3 recruitment of AMSH. Structure 2011, 19, 1149–1159.
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869.
- Kikuchi, K.; Ishii, N.; Asao, H.; Sugamura, K. Identification of AMSH-LP containing a Jab1/MPN domain metalloenzyme motif. Biochem. Biophys. Res. Commun. 2003, 306, 637–643.
- Davies, C.W.; Paul, L.N.; Kim, M.-I.; Das, C. Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: Nearly identical fold but different stability. J. Mol. Biol. 2011, 413, 416–429.
- Ren, G.-M.; Li, J.; Zhang, X.-C.; Wang, Y.; Xiao, Y.; Zhang, X.-Y.; Liu, X.; Zhang, W.; Ma, W.-B.; Zhang, J.; et al. Pharmacological targeting of NLRP3 deubiquitination for treatment of NLRP3-associated inflammatory diseases. Sci. Immunol. 2021, 6, eabe2933.
- Li, J.; Zhang, Y.; Da Silva Sil Dos Santos, B.; Wang, F.; Ma, Y.; Perez, C.; Yang, Y.; Peng, J.; Cohen, S.M.; Chou, T.-F.; et al. Epidithiodiketopiperazines inhibit protein degradation by targeting proteasome deubiquitinase Rpn11. Cell Chem. Biol. 2018, 25, 1350–1358.e1359.
- Song, Y.; Li, S.; Ray, A.; Das, D.S.; Qi, J.; Samur, M.K.; Tai, Y.T.; Munshi, N.; Carrasco, R.D.; Chauhan, D.; et al. Blockade of deubiquitylating enzyme Rpn11 triggers apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Oncogene 2017, 36, 5631–5638.
- Li, J.; Yakushi, T.; Parlati, F.; Mackinnon, A.L.; Perez, C.; Ma, Y.; Carter, K.P.; Colayco, S.; Magnuson, G.; Brown, B.; et al. Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat. Chem. Biol. 2017, 13, 486–493.
- Schlierf, A.; Altmann, E.; Quancard, J.; Jefferson, A.B.; Assenberg, R.; Renatus, M.; Jones, M.; Hassiepen, U.; Schaefer, M.; Kiffe, M.; et al. Targeted inhibition of the COP9 signalosome for treatment of cancer. Nat. Commun. 2016, 7, 13166.
- Liu, Y.; Liu, X.; Zhang, N.; Yin, M.; Dong, J.; Zeng, Q.; Mao, G.; Song, D.; Liu, L.; Deng, H. Berberine diminishes cancer cell PD-L1 expression and facilitates antitumor immunity inhibiting the deubiquitination activity of CSN5. Acta Pharm. Sin. B 2020, 10, 2299–2312.
- Dinarello, C.A. Biologic basis for interleukin-1 in disease. Blood 1996, 87, 2095–2147.
- Khare, S.; Dorfleutner, A.; Bryan, N.B.; Yun, C.; Radian, A.D.; de Almeida, L.; Rojanasakul, Y.; Stehlik, C. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 2012, 36, 464–476.
- Mikhael, J.R.; Dingli, D.; Roy, V.; Reeder, C.B.; Buadi, F.K.; Hayman, S.R.; Dispenzieri, A.; Fonseca, R.; Sher, T.; Kyle, R.A.; et al. Management of newly diagnosed symptomatic multiple myeloma: Updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin. Proc. 2013, 88, 360–376.
- Hadjiaggelidou, C.; Katodritou, E. Regulatory T-cells and multiple myeloma: Implications in tumor immune biology and treatment. J. Clin. Med. 2021, 10, 4588.
- Van der Linden, W.A.; Willems, L.I.; Shabaneh, T.B.; Li, N.; Ruben, M.; Florea, B.I.; van der Marel, G.A.; Kaiser, M.; Kisselev, A.F.; Overkleeft, H.S. Discovery of a potent and highly β1 specific proteasome inhibitor from a focused library of urea-containing peptide vinyl sulfones and peptide epoxyketones. Org. Biomol. Chem. 2012, 10, 181–194.
- Desvergne, A.; Genin, E.; Maréchal, X.; Gallastegui, N.; Dufau, L.; Richy, N.; Groll, M.; Vidal, J.; Reboud-Ravaux, M. Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome. J. Med. Chem. 2013, 56, 3367–3378.
- Kawamura, S.; Unno, Y.; List, A.; Mizuno, A.; Tanaka, M.; Sasaki, T.; Arisawa, M.; Asai, A.; Groll, M.; Shuto, S. Potent proteasome inhibitors derived from the unnatural cis-cyclopropane isomer of Belactosin A: Synthesis, biological activity, and mode of action. J. Med. Chem. 2013, 56, 3689–3700.
- Ozcan, S.; Kazi, A.; Marsilio, F.; Fang, B.; Guida, W.C.; Koomen, J.; Lawrence, H.R.; Sebti, S.M. Oxadiazole-isopropylamides as potent and noncovalent proteasome inhibitors. J. Med. Chem. 2013, 56, 3783–3805.
- Dimopoulos, M.A.; Richardson, P.G.; Moreau, P.; Anderson, K.C. Current treatment landscape for relapsed and/or refractory multiple myeloma. Nat. Rev. Clin. Oncol. 2015, 12, 42–54.
- Perez, C.; Li, J.; Parlati, F.; Rouffet, M.; Ma, Y.; Mackinnon, A.L.; Chou, T.-F.; Deshaies, R.J.; Cohen, S.M. Discovery of an inhibitor of the proteasome subunit Rpn11. J. Med. Chem. 2017, 60, 1343–1361.
- Lauinger, L.; Li, J.; Shostak, A.; Cemel, I.A.; Ha, N.; Zhang, Y.; Merkl, P.E.; Obermeyer, S.; Stankovic-Valentin, N.; Schafmeier, T.; et al. Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases. Nat. Chem. Biol. 2017, 13, 709–714.
- Li, B.; Wever, W.J.; Walsh, C.T.; Bowers, A.A. Dithiolopyrrolones: Biosynthesis, synthesis, and activity of a unique class of disulfide-containing antibiotics. Nat. Prod. Rep. 2014, 31, 905–923.
- Lee, M.-H.; Zhao, R.; Phan, L.; Yeung, S.-C.J. Roles of COP9 signalosome in cancer. Cell Cycle 2011, 10, 3057–3066.
- Lee, Y.H.; Judge, A.D.; Seo, D.; Kitade, M.; Gómez-Quiroz, L.E.; Ishikawa, T.; Andersen, J.B.; Kim, B.K.; Marquardt, J.U.; Raggi, C.; et al. Molecular targeting of CSN5 in human hepatocellular carcinoma: A mechanism of therapeutic response. Oncogene 2011, 30, 4175–4184.
- Altmann, E.; Erbel, P.; Renatus, M.; Schaefer, M.; Schlierf, A.; Druet, A.; Kieffer, L.; Sorge, M.; Pfister, K.; Hassiepen, U.; et al. Azaindoles as zinc-binding small-molecule inhibitors of the JAMM protease CSN5. Angew. Chem. 2017, 56, 1294–1297.
- Ni, W.-J.; Ding, H.-H.; Tang, L.-Q. Berberine as a promising anti-diabetic nephropathy drug: An analysis of its effects and mechanisms. Eur. J. Pharmacol. 2015, 760, 103–112.
- Liu, B.; Wang, G.; Yang, J.; Pan, X.; Yang, Z.; Zang, L. Berberine inhibits human hepatoma cell invasion without cytotoxicity in healthy hepatocytes. PLoS ONE 2011, 6, e21416.
- Chidambara Murthy, K.N.; Jayaprakasha, G.K.; Patil, B.S. The natural alkaloid berberine targets multiple pathways to induce cell death in cultured human colon cancer cells. Eur. J. Pharmacol. 2012, 688, 14–21.
This entry is offline, you can click here to edit this entry!
