Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
G protein-coupled receptors (GPCRs) are a large membrane protein family found in higher organisms, including the human body. GPCRs mediate cellular responses to diverse extracellular stimuli and thus control key physiological functions, which makes them important targets for drug design. Signaling by GPCRs is related to the structure and dynamics of these proteins, which are modulated by extrinsic ligands as well as by intracellular binding partners such as G proteins and arrestins.
- G protein-coupled receptors
- 19F-NMR
- membrane mimetics
- stable-isotope labeling
1. 19F-Nuclear Magnetic Resonance (NMR) with Observation of Extrinsic Probes Attached to G Protein-coupled Receptor (GPCR)
19F-NMR has long been used for studies of complex biological systems, since 19F has no natural background signals and displays high sensitivity toward changes in its microenvironment. As fluorine is not a natural component of proteins, it is essential to develop ever-improved methods to incorporate fluorine probes into GPCRs, either during expression or by post-translational chemical modification (Figure 1) [1][2][3][4][5].
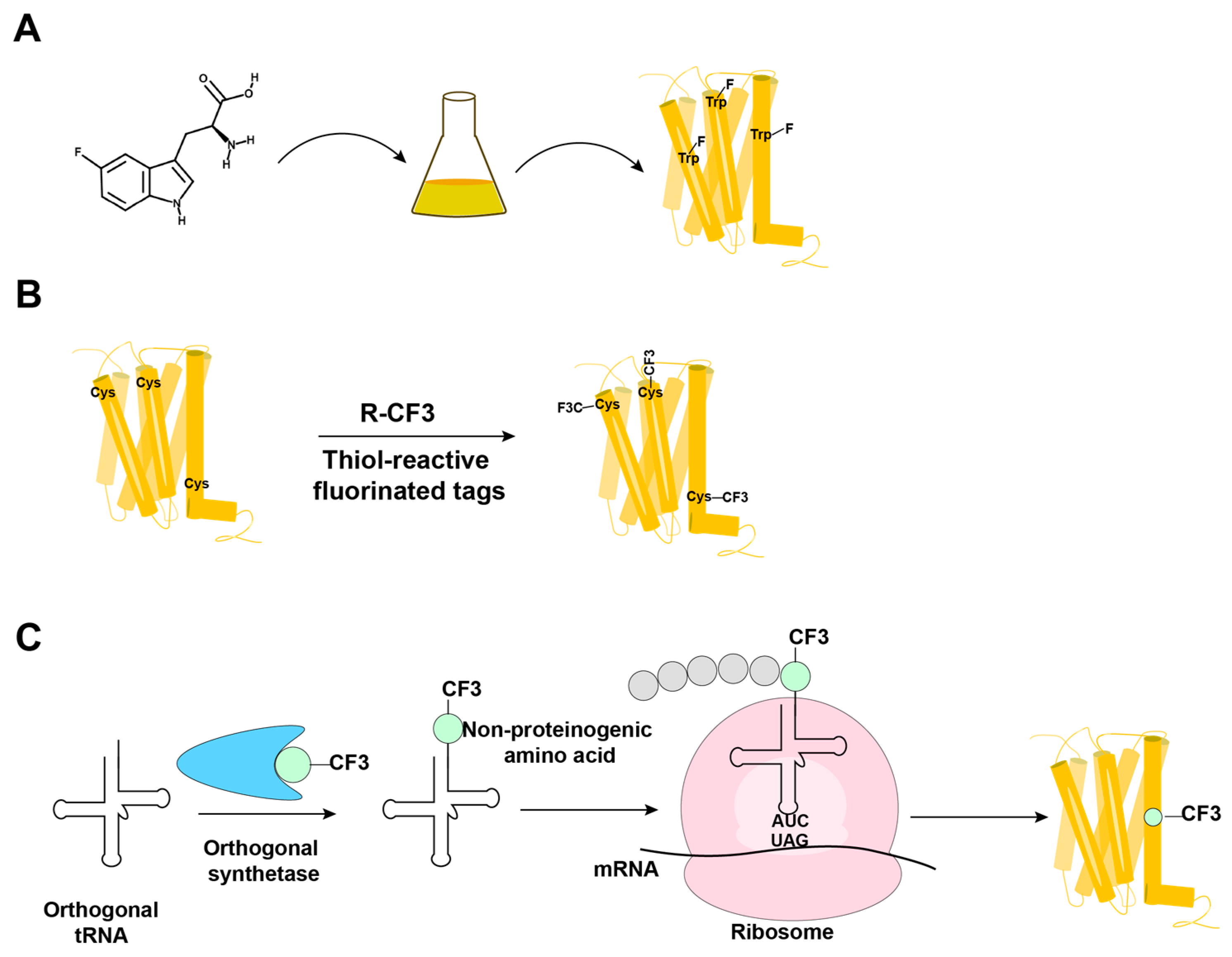
Figure 1. Overview of the methods in use for incorporation of 19F-NMR labels into GPCRs. (A) Biosynthetic incorporation by adding fluorinated amino acids, such as 5F-Trp, to the expression system; all Trp residues in the protein are then labeled with 19F. (B) Post-translational chemical modification by reacting the GPCR with thiol-reactive fluorinated tags; all reagent-accessible Cys residues are then labeled with the fluorinated tag. (C) Genetic labeling using an extrinsic orthogonal tRNA/aminoacyl-tRNA synthetase pair to incorporate non-proteinogenic 19F-labeled amino acids at positions defined by a TAG amber codon.
In biosynthetic incorporation, all residues of one amino acid type can be replaced by its fluorinated analogue, providing “amino-acid-specific 19F-labeling” (Figure 1A). Fluorinated amino acids are fed to the expression host by including a high concentration of the fluorinated amino acid in the growth medium (Figure 1A) [2][6][7]. This approach may be limited by the fact that high concentrations of 19F-containing amino acids can inhibit cell growth [8]. Induction of the amino acid auxotrophy in nonauxotrophic bacterial strains by shutting down selected amino-acid-specific biosynthesis pathways with specific inhibitors has also been used for 19F-labeling [9][10]. Tryptophan residues are highly present at the hydrophobic interfaces of protein–protein complexes, which makes fluorotryptophan attractive for NMR studies of membrane proteins [11][12]. The use of fluorinated indole as a fluorotryptophan precursor has been described as an inexpensive alternative for obtaining tryptophan-specific labeling [8].
Fluorine has been widely incorporated into proteins by post-translational chemical modification (Figure 1B). The most commonly employed method is cysteine-labeling, making use of the high nucleophilicity of the side chain sulfhydryl group [4]. Labels with CF3 groups are attractive because they yield strong signals that are not subject to large chemical shift anisotropy relaxation [5]. Examples are 3-bromo-1,1,1-trifluoroacetone (BTFA) [13] and 2-bromo-4-(trifluoromethyl)acetanilide (BTFMA), which react with the sulfhydryl group in a single step [14]. The conjugation of 2,2,2-trifluoroethanethiol (TET) to membrane proteins starts with sulfhydryl group activation by 4,4-dithiodipyridine (4-DPS), and a disulfide bond is formed in a second step [3][15][16][17][18]. Post-translational chemical modification can be applied to otherwise unlabeled proteins and regardless of the expression system used; high expression yields can thus be obtained, which is especially useful for GPCRs [3][15][16][17][18][19]. When using amino-acid-specific labeling, further sequence-specific assignments of 19F-resonances have been obtained by site-specific mutagenesis. For cysteine-rich GPCRs, individual assignments can therefore be very demanding. In 2015, Sušac et al. [17] reported the in-membrane chemical modification (IMCM) method, which makes use of the natural protection of most cysteines in the transmembrane helices by the membrane environment. Selective cysteine labeling on the receptor surface with minimal or no mutagenesis can thus be achieved.
The introduction of multiple 19F-labels within the same protein has been used to check on intramolecular distances related to the three-dimensional molecular structure [20][21].
In a genetic engineering approach, the site-specific incorporation of fluorinated amino acids is accomplished through using an orthogonal amber suppressor tRNA with a paired tRNA synthetase to insert the non-proteinogenic amino acid at positions defined by a TAG amber codon (Figure 1C) [1]. Fluorinated phenylalanine [22][23][24] and tyrosine [25] derivatives have been incorporated into proteins using this approach. Genetic labeling can be highly precise, but in improperly optimized expression systems, it may provide low yields of both the expression and incorporation of the fluorinated-amino acid [26]. Wang et al. [24] reported the genetic labeling of the cannabinoid receptor 1 (CB1) with the non-proteinogenic amino acid 3’-trifluoromenthyl-phenylalanine (mtfF) in the baculovirus expression system; this approach enabled studies of conformational transformations under the influence of ligands with variable efficacies [24].
2. NMR in Solution of GPCRs Using Stable-Isotope Labeling
Three different stable-isotope labeling strategies have primarily been used for studies of GPCRs: post-translational chemical labeling with 13C-labeled methyl groups, amino-acid-type selective labeling and uniform labeling. Post-translational chemical labeling of the reactive side chains of surface-accessible cysteine or lysine residues has been used in many NMR studies of GPCRs. 13C-isotope-labeled methyl probes can be chemically attached to the γ-SH moiety of cysteine side chains or the ε-NH2 groups of lysine side chains. 13C-formaldehyde has been used to label solvent-exposed lysines, yielding 13C-dimethyllysines as NMR probes [27][28][29]. 13C-methyl methanethiosulfonate (13C-MMTS) has been used to label solvent-exposed cysteines of GPCRs [30]. Unlike trifluoromethyl probes, which are usually attached to a single judiciously selected surface-accessible cysteine to avoid signal interference from other labeled residues [3][31][32][33][34], 13C-labeled methyl probes have often been used to label all surface-accessible (endogenous as well as non-endogenous) cysteines or lysines, and 2D 1H-13C correlation spectra were recorded to resolve multiple signals [35].
Limitations arise because the choice of stable-isotope probes for the chemical modification of amino acids side chains may influence the dynamics of GPCRs [33]. On principal grounds, post-translational chemical labeling normally only targets surface-accessible residues of GPCRs. In contrast, amino-acid-selective isotope labeling also targets the transmembrane region of the receptor. Examples of amino-acid-type selective isotope labeling include the use of [δ1-13CH3]-isoleucine [36][37], [ε-13CH3]-methionine [29][30][36][38][39][40][41][42][43][44][45][46], [15N]-valine [47][48] and [15N]-leucine [49]. An increased sensitivity was achieved by deuterating the α- and β- positions of methionine and leucine, using [2,3,3-2H, methyl-13C]-methionine [38][39][41][42] and [2,3,3-2H, 15N]-leucine [49] in the nutrient.
Uniform 15N-labeling of GPCRs has been achieved in E. coli for the rat neurotensin receptor 1 [50] and in Pichia pastoris for A2AAR [51][52][53] and the histamine H1 receptor [54]; the minimal medium contained [15N]-ammonium sulfate or [15N]-ammonium chloride as the only nitrogen sources. Since deuteration is mandatory for transverse relaxation-optimized spectroscopy (TROSY) studies of large macromolecular systems [55], it is essential that the expression systems used can produce partially or fully deuterated recombinant proteins. Uniform 2H, 15N-labeling was also achieved by expression of the β1-adrenergic receptor in Sf9 insect cells; the addition of 2H, 15N-labeled yeast extract to the insect cell medium allowed deuteration levels of >60% [56]. Eddy et al. [51][52][53] expressed uniformly 2H, 15N-labeled A2AAR with D2O-adapted Pichia pastoris in D2O growth media. All six tryptophan indole 15N-1H signals and eight of the eighteen glycine backbone 15N-1H NMR signals were resolved in the 2D [15N, 1H]-TROSY spectrum of A2AAR, and sequence-specific NMR assignments were obtained by single-residue amino acid replacements [51]. Drug-dependent local conformational changes in A2AAR could thus be observed, as illustrated for the toggle switch Trp2466.48 [51]. In addition to the natural tryptophans, extrinsic tryptophan residues were introduced into judiciously selected sites of the receptor by genetic engineering; these were then used as supplementary NMR probes for monitoring conformational changes of A2AAR [52][53].
Overall, in contrast to “probe methods”, uniform labeling can provide global information on a receptor.
3. GPCR–Ligand Interactions Studied by NMR Observation of the Ligand
NMR observation of bound and free ligands can provide unique insights into the biophysical properties and biological functions of GPCRs. Specifically, in addition to providing data on the influence of bound ligands on GPCR, NMR spectroscopy is uniquely powerful in detecting weak binding [57][58][59]. Measurements of the chemical shifts, line widths, and relaxation times of free and bound ligands are all informative on ligand binding events. Depending on the time scale of the GPCR–ligand interactions, different approaches are used [60].
For studies of weak binding with a rapid ligand exchange, a large arsenal of experiments based on observation of the modulation of the NMR signal of the free ligand through exchange with the bound ligand is available [61][62][63][64]. Among these, the transfer NOE (trNOE) stands out by the fact that information on the structure of the bound ligand can be obtained. For example, in studies of the peptide ligand dynorphin interacting with the kappa opioid receptor (KOR) [65], 1H and 15N chemical shift variations for the free ligand indicated that the free peptide is in fast exchange with the bound peptide. The receptor–peptide interaction was within the range that allowed the determination of a conformation of the KOR-bound dynorphin via the trNOE method.
This entry is adapted from the peer-reviewed paper 10.3390/molecules27092658
References
- Wang, L.; Brock, A.; Herberich, B.; Schultz, P.G. Expanding the genetic code of Escherichia coli. Science 2001, 292, 498–500.
- Frieden, C.; Hoeltzli, S.D.; Bann, J.G. The preparation of 19F-labeled proteins for NMR studies. Methods Enzymol. 2004, 380, 400–415.
- Liu, J.J.; Horst, R.; Katritch, V.; Stevens, R.C.; Wüthrich, K. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 2012, 335, 1106–1110.
- Kitevski-LeBlanc, J.L.; Prosser, R.S. Current applications of 19F-NMR to studies of protein structure and dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2012, 62, 1–33.
- Didenko, T.; Liu, J.J.; Horst, R.; Stevens, R.C.; Wüthrich, K. Fluorine-19 NMR of integral membrane proteins illustrated with studies of GPCRs. Curr. Opin. Struct. Biol. 2013, 23, 740–747.
- Lu, P.; Jarema, M.; Mosser, K.; Daniel, W.E. Lac repressor: 3-fluorotyrosine substitution for nuclear magnetic resonance studies. Proc. Natl. Acad. Sci. USA 1976, 73, 3471–3475.
- Hou, Y.; Hu, W.; Li, X.; Skinner, J.J.; Liu, D.; Wüthrich, K. Solvent-accessibility of discrete residue positions in the polypeptide hormone glucagon by 19F-NMR observation of 4-fluorophenylalanine. J. Biomol. NMR 2017, 68, 1–6.
- Crowley, P.B.; Kyne, C.; Monteith, W.B. Simple and inexpensive incorporation of 19F-tryptophan for protein NMR spectroscopy. Chem. Commun. 2012, 48, 10681–10683.
- Steinrucken, H.C.; Amrhein, N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 1980, 94, 1207–1212.
- Arntson, K.E.; Pomerantz, W.C. Protein-observed fluorine NMR: A bioorthogonal approach for small molecule discovery. J. Med. Chem. 2016, 59, 5158–5171.
- Bogan, A.A.; Thorn, K.S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998, 280, 1–9.
- Picard, L.P.; Prosser, R.S. Advances in the study of GPCRs by 19F-NMR. Curr. Opin. Struct. Biol. 2021, 69, 169–176.
- Hellmich, U.A.; Pfleger, N.; Glaubitz, C. 19F-MAS NMR on proteorhodopsin: Enhanced protocol for site-specific labeling for general application to membrane proteins. Photochem. Photobiol. 2009, 85, 535–539.
- Manglik, A.; Kim, T.H.; Masureel, M.; Altenbach, C.; Yang, Z.; Hilger, D.; Lerch, M.T.; Kobilka, T.S.; Thian, F.S.; Hubbell, W.L.; et al. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell 2015, 161, 1101–1111.
- Horst, R.; Liu, J.J.; Stevens, R.C.; Wüthrich, K. β2-adrenergic receptor activation by agonists studied with 19F-NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2013, 52, 10762–10765.
- Ye, L.; Larda, S.T.; Frank Li, Y.F.; Manglik, A.; Prosser, R.S. A comparison of chemical shift sensitivity of trifluoromethyl tags: Optimizing resolution in 19F-NMR studies of proteins. J. Biomol. NMR 2015, 62, 97–103.
- Sušac, L.; O’Connor, C.; Stevens, R.C.; Wüthrich, K. In-membrane chemical modification (IMCM) for site-specific chromophore labeling of GPCRs. Angew. Chem. Int. Ed. Engl. 2015, 54, 15246–15249.
- Wang, H.; Hu, W.; Liu, D.; Wüthrich, K. Design and preparation of the class B G protein-coupled receptors GLP-1R and GCGR for 19F-NMR studies in solution. FEBS J. 2021, 288, 4053–4063.
- Chung, K.Y.; Kim, T.H.; Manglik, A.; Alvares, R.; Kobilka, B.K.; Prosser, R.S. Role of detergents in conformational exchange of a G protein-coupled receptor. J. Biol. Chem. 2012, 287, 36305–36311.
- Orton, H.W.; Qianzhu, H.; Abdelkader, E.H.; Habel, E.I.; Tan, Y.J.; Frkic, R.L.; Jackson, C.J.; Huber, T.; Otting, G. Through-space scalar 19F–19F couplings between fluorinated noncanonical amino acids for the detection of specific contacts in proteins. J. Am. Chem. Soc. 2021, 143, 19587–19598.
- Loewen, M.C.; Klein-Seetharaman, J.; Getmanova, E.V.; Reeves, P.J.; Schwalbe, H.; Khorana, H.G. Solution 19F nuclear overhauser effects in structural studies of the cytoplasmic domain of mammalian rhodopsin. Proc. Natl. Acad. Sci. USA 2001, 98, 4888–4892.
- Jackson, J.C.; Hammill, J.T.; Mehl, R.A. Site-specific incorporation of a 19F-amino acid into proteins as an NMR probe for characterizing protein structure and reactivity. J. Am. Chem. Soc. 2007, 129, 1160–1166.
- Cellitti, S.E.; Jones, D.H.; Lagpacan, L.; Hao, X.; Zhang, Q.; Hu, H.; Brittain, S.M.; Brinker, A.; Caldwell, J.; Bursulaya, B.; et al. In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 2008, 130, 9268–9281.
- Wang, X.; Liu, D.; Shen, L.; Li, F.; Li, Y.; Yang, L.; Xu, T.; Tao, H.; Yao, D.; Wu, L.; et al. A genetically encoded F-19 NMR probe reveals the allosteric modulation mechanism of cannabinoid receptor 1. J. Am. Chem. Soc. 2021, 143, 16320–16325.
- Minnihan, E.C.; Young, D.D.; Schultz, P.G.; Stubbe, J. Incorporation of fluorotyrosines into ribonucleotide reductase using an evolved, polyspecific aminoacyl-tRNA synthetase. J. Am. Chem. Soc. 2011, 133, 15942–15945.
- Young, T.S.; Ahmad, I.; Yin, J.A.; Schultz, P.G. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010, 395, 361–374.
- Bokoch, M.P.; Zou, Y.; Rasmussen, S.G.; Liu, C.W.; Nygaard, R.; Rosenbaum, D.M.; Fung, J.J.; Choi, H.J.; Thian, F.S.; Kobilka, T.S.; et al. Ligand-specific regulation of the extracellular surface of a G protein-coupled receptor. Nature 2010, 463, 108–112.
- Sounier, R.; Mas, C.; Steyaert, J.; Laeremans, T.; Manglik, A.; Huang, W.; Kobilka, B.K.; Déméné, H.; Granier, S. Propagation of conformational changes during μ-opioid receptor activation. Nature 2015, 524, 375–378.
- Cong, X.; Maurel, D.; Demene, H.; Vasiliauskaite-Brooks, I.; Hagelberger, J.; Peysson, F.; Saint-Paul, J.; Golebiowski, J.; Granier, S.; Sounier, R. Molecular insights into the biased signaling mechanism of the μ-opioid receptor. Mol. Cell 2021, 81, 4165–4175.e6.
- Goba, I.; Goricanec, D.; Schum, D.; Hillenbrand, M.; Pluckthun, A.; Hagn, F. Probing the conformation states of neurotensin receptor 1 variants by NMR site-directed methyl labeling. Chembiochem 2021, 22, 139–146.
- Klein-Seetharaman, J.; Getmanova, E.V.; Loewen, M.C.; Reeves, P.J.; Khorana, H.G. NMR spectroscopy in studies of light-induced structural changes in mammalian rhodopsin: Applicability of solution 19F-NMR. Proc. Natl. Acad. Sci. USA 1999, 96, 13744–13749.
- Kim, T.H.; Chung, K.Y.; Manglik, A.; Hansen, A.L.; Dror, R.O.; Mildorf, T.J.; Shaw, D.E.; Kobilka, B.K.; Prosser, R.S. The role of ligands on the equilibria between functional states of a G protein-coupled receptor. J. Am. Chem. Soc. 2013, 135, 9465–9474.
- Prosser, R.S.; Ye, L.; Pandey, A.; Orazietti, A. Activation processes in ligand-activated G protein-coupled receptors: A case study of the adenosine A2A receptor. Bioessays 2017, 39, 1700072.
- Frei, J.N.; Broadhurst, R.W.; Bostock, M.J.; Solt, A.; Jones, A.J.Y.; Gabriel, F.; Tandale, A.; Shrestha, B.; Nietlispach, D. Conformational plasticity of ligand-bound and ternary GPCR complexes studied by 19F-NMR of the β1-adrenergic receptor. Nat. Commun. 2020, 11, 669.
- Ma, X.; Hu, Y.; Batebi, H.; Heng, J.; Xu, J.; Liu, X.; Niu, X.; Li, H.; Hildebrand, P.W.; Jin, C.; et al. Analysis of β2AR-Gs and β2AR-Gi complex formation by NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 23096–23105.
- Casiraghi, M.; Damian, M.; Lescop, E.; Point, E.; Moncoq, K.; Morellet, N.; Levy, D.; Marie, J.; Guittet, E.; Baneres, J.L.; et al. Functional modulation of a G protein-coupled receptor conformational landscape in a lipid bilayer. J. Am. Chem. Soc. 2016, 138, 11170–11175.
- Clark, L.D.; Dikiy, I.; Chapman, K.; Rödström, K.E.; Aramini, J.; LeVine, M.V.; Khelashvili, G.; Rasmussen, S.G.; Gardner, K.H.; Rosenbaum, D.M. Ligand modulation of sidechain dynamics in a wild-type human GPCR. Elife 2017, 6, e28505.
- Mizumura, T.; Kondo, K.; Kurita, M.; Kofuku, Y.; Natsume, M.; Imai, S.; Shiraishi, Y.; Ueda, T.; Shimada, I. Activation of adenosine A2A receptor by lipids from docosahexaenoic acid revealed by NMR. Sci. Adv. 2020, 6, eaay8544.
- Kofuku, Y.; Ueda, T.; Okude, J.; Shiraishi, Y.; Kondo, K.; Maeda, M.; Tsujishita, H.; Shimada, I. Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun. 2012, 3, 1045.
- Nygaard, R.; Zou, Y.; Dror, R.O.; Mildorf, T.J.; Arlow, D.H.; Manglik, A.; Pan, A.C.; Liu, C.W.; Fung, J.J.; Bokoch, M.P.; et al. The dynamic process of β2-adrenergic receptor activation. Cell 2013, 152, 532–542.
- Kofuku, Y.; Ueda, T.; Okude, J.; Shiraishi, Y.; Kondo, K.; Mizumura, T.; Suzuki, S.; Shimada, I. Functional dynamics of deuterated β2-adrenergic receptor in lipid bilayers revealed by NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2014, 53, 13376–13379.
- Okude, J.; Ueda, T.; Kofuku, Y.; Sato, M.; Nobuyama, N.; Kondo, K.; Shiraishi, Y.; Mizumura, T.; Onishi, K.; Natsume, M.; et al. Identification of a conformational equilibrium that determines the efficacy and functional selectivity of the μ-opioid receptor. Angew. Chem. Int. Ed. Engl. 2015, 54, 15771–15776.
- Solt, A.S.; Bostock, M.J.; Shrestha, B.; Kumar, P.; Warne, T.; Tate, C.G.; Nietlispach, D. Insight into partial agonism by observing multiple equilibria for ligand-bound and Gs-mimetic nanobody-bound β1-adrenergic receptor. Nat. Commun. 2017, 8, 1795.
- Bumbak, F.; Keen, A.C.; Gunn, N.J.; Gooley, P.R.; Bathgate, R.A.D.; Scott, D.J. Optimization and 13CH3 methionine labeling of a signaling competent neurotensin receptor 1 variant for NMR studies. Biochim. Biophys. Acta 2018, 1860, 1372–1383.
- Xu, J.; Hu, Y.; Kaindl, J.; Risel, P.; Hübner, H.; Maeda, S.; Niu, X.; Li, H.; Gmeiner, P.; Jin, C.; et al. Conformational complexity and dynamics in a muscarinic receptor revealed by NMR spectroscopy. Mol. Cell 2019, 75, 53–65.e7.
- Rößler, P.; Mayer, D.; Tsai, C.J.; Veprintsev, D.B.; Schertler, G.F.X.; Gossert, A.D. GPCR activation states induced by nanobodies and Mini-G proteins compared by NMR spectroscopy. Molecules 2020, 25, 5984.
- Isogai, S.; Deupi, X.; Opitz, C.; Heydenreich, F.M.; Tsai, C.J.; Brueckner, F.; Schertler, G.F.; Veprintsev, D.B.; Grzesiek, S. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 2016, 530, 237–241.
- Grahl, A.; Abiko, L.A.; Isogai, S.; Sharpe, T.; Grzesiek, S. A high-resolution description of β1-adrenergic receptor functional dynamics and allosteric coupling from backbone NMR. Nat. Commun. 2020, 11, 2216.
- Imai, S.; Yokomizo, T.; Kofuku, Y.; Shiraishi, Y.; Ueda, T.; Shimada, I. Structural equilibrium underlying ligand-dependent activation of β2-adrenoreceptor. Nat. Chem. Biol. 2020, 16, 430–439.
- Nasr, M.L.; Baptista, D.; Strauss, M.; Sun, Z.J.; Grigoriu, S.; Huser, S.; Pluckthun, A.; Hagn, F.; Walz, T.; Hogle, J.M.; et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 2017, 14, 49–52.
- Eddy, M.T.; Lee, M.Y.; Gao, Z.G.; White, K.L.; Didenko, T.; Horst, R.; Audet, M.; Stanczak, P.; McClary, K.M.; Han, G.W.; et al. Allosteric coupling of drug binding and intracellular signaling in the A2A adenosine receptor. Cell 2018, 172, 68–80.e12.
- Eddy, M.T.; Gao, Z.G.; Mannes, P.; Patel, N.; Jacobson, K.A.; Katritch, V.; Stevens, R.C.; Wüthrich, K. Extrinsic tryptophans as NMR probes of allosteric coupling in membrane proteins: Application to the A2A adenosine receptor. J. Am. Chem. Soc. 2018, 140, 8228–8235.
- Eddy, M.T.; Martin, B.T.; Wüthrich, K. A2A adenosine receptor partial agonism related to structural rearrangements in an activation microswitch. Structure 2021, 29, 170–176.e3.
- Mulry, E.; Ray, A.P.; Eddy, M.T. Production of a human histamine receptor for NMR spectroscopy in aqueous solutions. Biomolecules 2021, 11, 632.
- Pervushin, K.; Riek, R.; Wider, G.; Wüthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 1997, 94, 12366–12371.
- Opitz, C.; Isogai, S.; Grzesiek, S. An economic approach to efficient isotope labeling in insect cells using homemade 15N-, 13C- and 2H-labeled yeast extracts. J. Biomol. NMR 2015, 62, 373–385.
- Bemis, G.W.; Murcko, M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996, 39, 2887–2893.
- Fejzo, J.; Lepre, C.A.; Peng, J.W.; Bemis, G.W.; Ajay; Murcko, M.A.; Moore, J.M. The SHAPES strategy: An NMR-based approach for lead generation in drug discovery. Chem. Biol. 1999, 6, 755–769.
- Buratto, R.; Mammoli, D.; Chiarparin, E.; Williams, G.; Bodenhausen, G. Exploring weak ligand–protein interactions by long-lived NMR states: Improved contrast in fragment-based drug screening. Angew. Chem. Int. Ed. 2014, 53, 11376–11380.
- Millet, O.; Loria, J.P.; Kroenke, C.D.; Pons, M.; Palmer, A.G. The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J. Am. Chem. Soc. 2000, 122, 2867–2877.
- Chen, A.; Shapiro, M.J. NOE pumping: A novel NMR technique for identification of compounds with binding affinity to macromolecules. J. Am. Chem. Soc. 1998, 120, 10258–10259.
- Klein, J.; Meinecke, R.; Mayer, M.; Meyer, B. Detecting binding affinity to immobilized receptor proteins in compound libraries by HR-MAS STD NMR. J. Am. Chem. Soc. 1999, 121, 5336–5337.
- Mayer, M.; Meyer, B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. 1999, 38, 1784–1788.
- Dalvit, C.; Pevarello, P.; Tatò, M.; Veronesi, M.; Vulpetti, A.; Sundström, M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR 2000, 18, 65–68.
- O’Connor, C.; White, K.L.; Doncescu, N.; Didenko, T.; Roth, B.L.; Czaplicki, G.; Stevens, R.C.; Wüthrich, K.; Milon, A. NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc. Natl. Acad. Sci. USA 2015, 112, 11852–11857.
This entry is offline, you can click here to edit this entry!
