Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Unlike herbaceous plants, woody plants undergo volumetric growth (a.k.a. secondary growth) through wood formation, during which the secondary xylem (i.e., wood) differentiates from the vascular cambium. Wood is the most abundant biomass on Earth and, by absorbing atmospheric carbon dioxide, functions as one of the largest carbon sinks. As a sustainable and eco-friendly energy source, lignocellulosic biomass can help address environmental pollution and the global climate crisis.
- woody plant
- biomass
- secondary growth
- wood formation
- cambium
- xylem
1. Introduction
Land plants can be divided into two main groups: vascular and non-vascular plants. The vascular system is one of the key factors that enabled plants to successfully settle on land about 470 million years ago. The vascular system not only transports water, nutrients, and signals throughout the plant but also serves as mechanical support that maintains the plant’s vertical growth, increasing access to sunlight. The secondary xylem consists of vessel, fiber, and tracheid cells, with a thick secondary cell wall (SCW) composed mainly of cellulose, hemicellulose, and lignin [1][2][3]. The secondary xylem is derived from the vascular cambium, which is a cylindrical secondary meristem. Wood formation is achieved through a series of cascading processes that include: xylem mother cell specification by vascular cambium cell division, the differentiation of these cells into xylem cells followed by cell expansion, secondary cell wall deposition, pit formation, and programmed cell death (Figure 1). Each step is elaborately coordinated by factors such as hormones, signal peptides, and transcription factors (TFs). Recently, due to the global climate crisis, interest in sustainable energy development using eco-friendly and renewable biomass is increasing. Woody biomass produced from wood formation processes offers an economic and sustainable feedstock for bioenergy production.
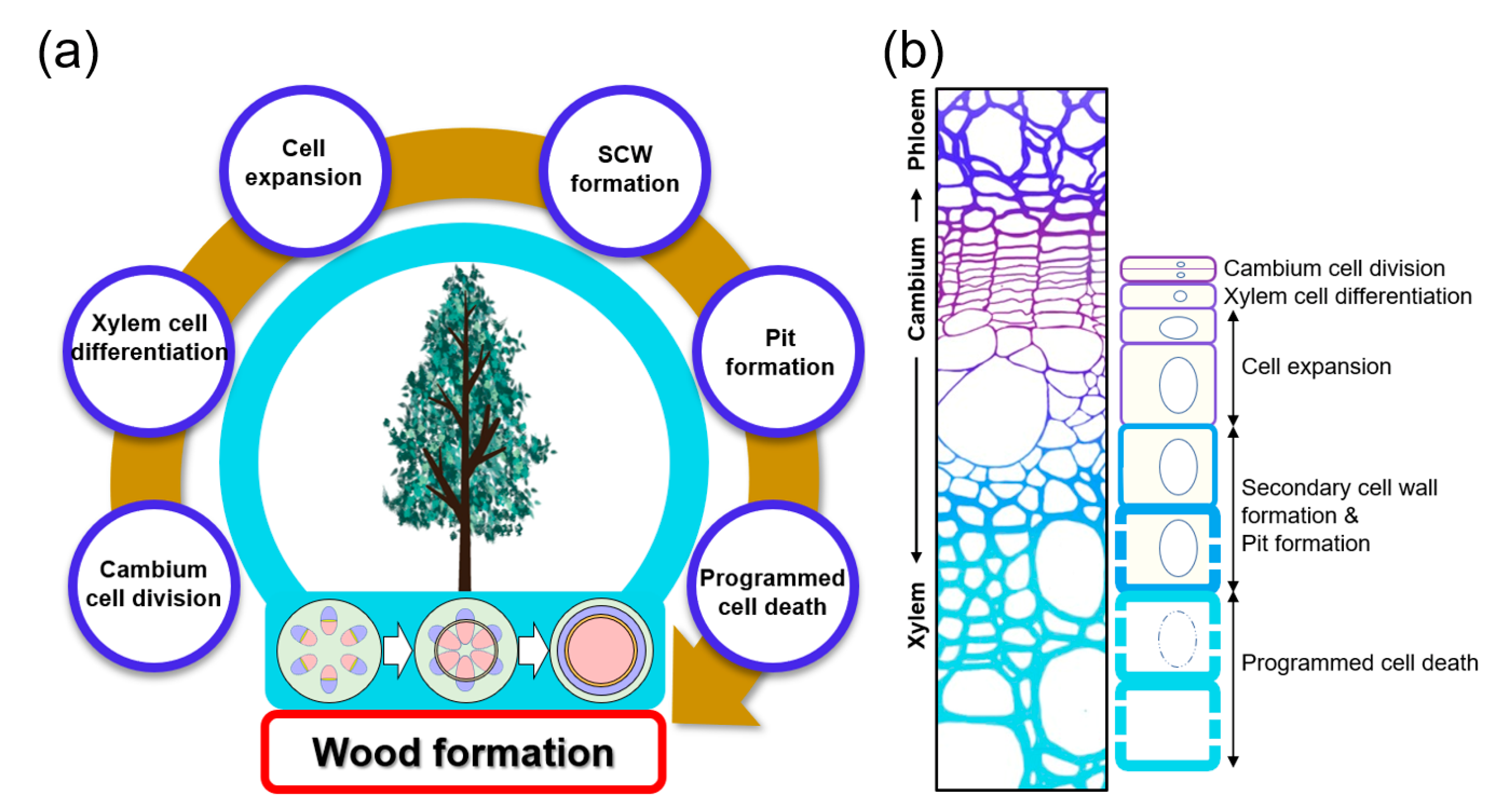
Figure 1. Wood formation in plants. (a) Simplified process of wood formation. Wood formation is initiated through cell divisions within the cylindrical vascular cambium layer, formed from the procambium. Then xylem cell differentiation, cell expansion, secondary cell wall (SCW) and pit formation, and programmed cell death (PCD) follow. Vascular cambium formation from procambium is shown in the stem cross-sections from below a tree: xylem (red), phloem (blue), and the cambium (yellow). (b) Stem cross-section and cell diagram for each stage of wood formation. The xylem is formed through the vascular cambium cell division and xylem cell differentiation, cell expansion, secondary cell wall formation and pit formation, and programmed cell death.
2. Vascular Cambium Cell Development and Xylem Cell Differentiation: Initiating Wood Formation
The growth and development of woody plants can be largely divided into primary and secondary phases. Primary growth occurs in the shoot apical meristem (SAM) within the ground and root apical meristem (RAM) in the basement. In the apical region of this meristem tissue, the procambium differentiates into primary phloem and primary xylem that comprise the primary vascular bundle. As the procambium expands to the interfascicular region, it develops into a ring to form vascular cambium [4]. Secondary growth refers to the production of wood by the vascular cambium. Vascular cambium divides to produce daughter cells, with secondary xylem formed toward the center and secondary phloem on the outside of the plant. Xylem mother cells can differentiate into several types of daughter cells. However, it is unknown how the cambium develops and how cell fate is determined via the cambium to the secondary xylem or secondary phloem.
Recently, considerable progress has been made in understanding the molecular mechanisms of cambium formation and development, based on research conducted in model plants such as Arabidopsis and poplar. These studies have revealed that cambium development is regulated by hormones, TFs, and signal peptides [5][6][7][8] (Figure 2a). A series of studies on the functional characteristics of Arabidopsis and poplar mutant plants have revealed genes that play an important role in the development of vascular cambium.
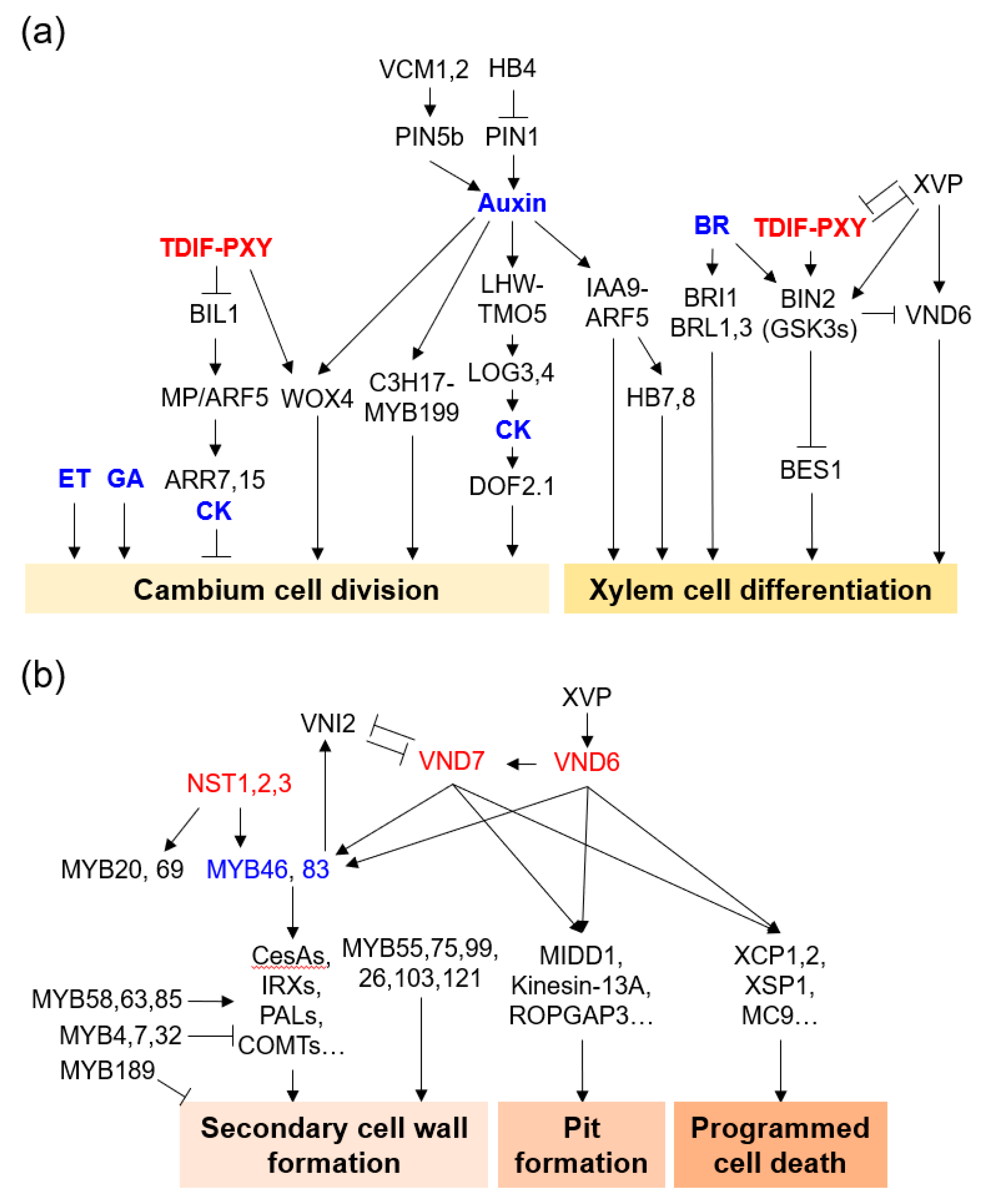
Figure 2. Molecular regulatory network of xylem cell formation. (a) Vascular cambium cell division and xylem cell differentiation. (b) Secondary cell wall formation, pit formation, and programmed cell death in xylem cell formation. Representative genes were depicted with hormones (blue). Genes with essential roles are described in the text.
2.1. Regulation by Plant Hormones
Hormones control various functions such as plant growth, development, and stress resistance [9]. Many studies have reported that vascular cambium activity is regulated by several hormones, including auxins, cytokinins, ethylene, and brassinosteroids.
Auxins are hormones that function in plant growth and development and affect cellular processes such as cell division, expansion, and differentiation [10][11]. Auxins are highly expressed in the cambium cell layer and play an important role in vascular cambium initiation and development [12][13][14][15][16]. PtoIAA9 and AUX/IAA in Populus tomentosa interact with AUXIN RESPONSE FACTOR 5 (PtoARF5) to regulate vascular cambium cell division and secondary xylem development. Auxin signaling maximum leads to direct activation of CLASS III HOMEODOMAIN-LEUCINE ZIPPER III (HD-ZIP III) TFs and changes in cell type-specific transcriptomes that define xylem cell identities [16][17]. The PtoIAA9-PtoARF5 module can bind to the promoter of the HD-ZIP III genes PtoHB7 and PtoHB8, which are involved in secondary xylem formation [18]. When the poplar HD-ZIP III gene PtrHB4 was upregulated, cambium development was induced by enhanced expression of PtrPIN1 [19]. The auxin-responsive module PaC3H17-PaMYB199 is associated with cambium cell division in poplar stems [20]. Knockdown mutants of two Populus MADS-box genes (VCM1 and VCM2), which are modulators of auxin homeostasis specifically expressed in the vascular cambium, enhanced vascular cambium proliferation activity and subsequent xylem differentiation [21].
Cytokinins (CKs) play important roles in vascular cambium development together with auxins. Poplar cytokinin receptor genes HISTIDINE KINASE 3 (PtHK3a) and PtHK3b are expressed at high levels in dividing cambium cells [22]. Inhibition of CK signaling by expression of Arabidopsis AtCKX2 under the promoter of the birch CRE1 gene reduced the number of cambium cells, whereas an increase in CK biosynthesis by expression of Arabidopsis ISOPENTENYL TRANSFERASE 7 (IPT7) increased cambium cell division in transgenic poplars [14][22]. Auxin-regulated LONESOME HIGHWAY (LHW) and TARGET OF MONOPTEROS5 (TMO5)/TMO5-LIKE1 (T5L1) directly up-regulated the CK biosynthesis genes LONELY GUY3 (LOG3) and LOG4, which are involved in the activation of vascular cell division [23]. AT2G28510/DOF2.1, which is a CK-dependent downstream target gene of LHW-TMO5/T5L1, controls vascular cell proliferation [24].
Brassinosteroids (BRs), ethylene (ET), and gibberellins (GAs) also promote vascular cambium division and secondary growth in trees. Mutations of both BRI-LIKE 1 (BRL1) and BRL3, which are Arabidopsis vascular-specific BR receptors, resulted in reduced xylem formation and increased phloem development [25]. BRI1-EMS SUPPRESSOR 1 (BES1) was shown to be involved in xylem differentiation downstream of TDIF-TDR-GSK3s signaling [26]. Similarly, inhibition of BR synthesis resulted in decreased secondary xylem differentiation and SCW biosynthesis, whereas increased BR levels increased secondary growth in poplar [27]. Exogenous BR treatment or genetic complementation of the BR biosynthesis DWARF gene in BR-biosynthetic-mutant tomato with retardation of xylem development resulted in a complete recovery of xylem cell formation [28]. In contrast, overexpression of GLYCOGEN SYNTHASE KINASE 3 (SlGSK3) or CRISPR/Cas9 knockout of BRASSINOSTEROID-INSENSITIVE 1 (SlBRI1) to block BR signaling resulted in severely defective xylem differentiation and secondary growth in tomato [28]. The tonoplast membrane-localized auxin efflux carrier WALLS ARE THIN1 (SlWAT1) is directly activated by SlBRL1/2 in xylem precursor cells. Transposable element (TE)-mediated loss-of-function allele Slwat1-copi resulted in defects in secondary xylem development, with a reduced vessel element number in tomato. Secondary xylem formation of Slwat1-copi was completely recovered by genetic complementation of WAT1 function [29]. Cambium division was increased in ET-overproducing poplar, whereas it was decreased in ET-insensitive poplar [30]. acs7-d, an ET overproducing Arabidopsis mutant, showed enhanced cambial activity and reduced fiber cell wall development [31]. Transgenic poplar overexpressing the GIBBERELLIN 20 OXIDASE 1 (GA20ox1) gene, which is involved in GA biosynthesis, was characterized by GA overproduction and cambium proliferation [32][33][34]. GA also induced vascular cambium differentiation and lignification when expressed downstream of WOX14 in Arabidopsis stems [35][36].
2.2. Regulation by Transcription Factors and Signal Peptides
Regulation via TFs and signal peptides is essential for vascular cambium development and xylem differentiation. CLAVATA3/EMBRYO SURROUNDING REGION (CLE) family peptides are 12–13 amino acids with two hydroxylated proline residues after processing and post-translational modifications [37]. CLE41/44 (a.k.a. TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR [TDIF]) is secreted by developing phloem cells and binds to the cambium cell receptor PHLOEM INTERCALATED WITH XYLEM (PXY) [38]. The TDIF-PXY module plays an important role in maintaining the cambium cell population via cell division and inhibiting xylem cell differentiation [39][40]. WUSCHEL HOMEOBOX RELATED 4 (WOX4), a downstream transcriptional regulator of the TDIF-PXY module, is specifically expressed in the cambium region, and is involved in the regulation of cambium activity [38][39][40][41][42]. Ectopic expression of PttCLE41b and other related PttCLE41-like genes causes a dwarf phenotype with loss of cell division orientation and defects in the patterning of vascular tissues [42]. PtrCLE20 is expressed in xylem tissues and suppresses cambium activity in poplar [43]. Populus WOX4, PttWOX4, is specifically expressed in the cambial region during the growing season only. PttWOX4a/b RNAi transgenic poplar showed reduced vascular cambium width and secondary growth [42]. Auxin-dependent WOX4 regulation is achieved in xylem precursor cells with auxin maxima, which promote vascular cambium cell division in a non-cell-autonomous manner [16]. GLYCOGEN SYNTHASE KINASE3/SHAGGY-LIKE KINASE proteins (GSK3s), including ARABIDOPSIS BRASSINOSTEROID-INSENSITIVE2 (BIN2), BIN2-LIKE1 (BIL1), and BIL2 are downstream of the TDIF-PXY module and inhibit xylem differentiation via the suppression of BRI1–EMS–SUPPRESSOR1 (BES1) and BRASSINAZOLE RESISTANT 1 (BZR1) [26][44]. BIL1 is a key mediator linking the TDIF-PXY module with auxin-CK signaling for the maintenance of cambium activity [45]. BIL1 phosphorylates and activates MONOPTEROS/AUXIN RESPONSE FACTOR 5 (MP/ARF5), and activated MP promotes the expression of the CK signaling negative regulators ARABIDOPSIS RESPONSE REGULATOR 7 (ARR7) and ARR15, which suppress cambium cell division.
HD-ZIP III and NAC (NAM, ATAF and CUC) TFs have been reported to play important roles in wood formation. PopREVOLUTA, the closest poplar homolog gene to Arabidopsis REVOLUTA (REV), has been reported to be involved in vascular cambium initiation [46]. Poplar PtrHB5 and PtrHB7, which are the closest homologs of Arabidopsis CORONA and AtHB8, induce cambium activity and xylem differentiation during secondary growth [47][48]. PtrHB7 is preferentially expressed in cambium tissues and is a direct target of the PtrIAA9-PtrARF5 module, inducing cambium activity and xylem differentiation [18][47][48]. PtrHB4 is specifically expressed in shoot tips and in the early developmental stages of vascular tissue; PtrHB4-SRDX (PtrHB4 repressor) transgenic poplar showed defects in the secondary vascular system due to failure of interfascicular cambium formation [19].
The NAC TF family have a highly conserved N-terminal NAC domain, which is associated with nuclear localization, DNA binding, and homodimer and/or heterodimer formation with other NAC proteins [49]. Among the NAC TFs in Arabidopsis, VASCULAR-RELATED NAC DOMAINs (VNDs) are master regulators of xylem differentiation [50][51]. NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1 and 3 (NST1 and 3) are involved in stem fiber differentiation [52][53]. VND1, VND2, and VND3 contribute to xylem vessel formation during seedling development [54]. Poplar VNDs and NST1/3 are also involved in xylem differentiation [55][56][57]. VND6 and VND7, in particular, have been studied as master switches of xylem differentiation in metaxylem and protoxylem cells, respectively [50][58][59]. VND6 and VND7 upregulate xylem vessel cell differentiation-related genes such as SCW biosynthesis genes and programmed cell death (PCD) genes [58][60][61][62]. VND6 and VND7 directly regulate MYB46 and MYB83, which are master regulators of SCW biosynthesis [63][64]. XYLEM DIFFERENTIATION AND ALTERED VASCULAR PATTERNING (XVP) is a negative regulator of the TDIF-PXY module and fine-tunes TDIF signaling for xylem differentiation via interacting with the PXY-BAK1 (BRI1-ASSOCIATED KINASE 1) receptor complex [65].
This entry is adapted from the peer-reviewed paper 10.3390/genes13071181
References
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531.
- Morris, H.; Plavcova, L.; Cvecko, P.; Fichtler, E.; Gillingham, M.; Martínez-Cabrera, H.; McGlinn, D.; Wheeler, E.; Zheng, J.; Ziemińska, K.; et al. A global analysis of parenchyma tissue fractions in secondary xylem of seed plants. New Phytol. 2015, 209, 1553–1565.
- Rajput, K.S.; Gondaliya, A.D.; Lekhak, M.M.; Yadav, S.R. Structure and Ontogeny of Intraxylary Secondary Xylem and Phloem Development by the Internal Vascular Cambium in Campsis radicans (L.) Seem. (Bignoniaceae). J. Plant Growth Regul. 2018, 37, 755–767.
- Nieminen, K.; Blomster, T.; Helariutta, Y.; Mähönen, A.P. Vascular Cambium Development. Arab. Book 2015, 13, e0177.
- Miyashima, S.; Sebastian, J.; Lee, J.Y.; Helariutta, Y. Stem cell function during plant vascular development. EMBO J. 2013, 32, 178–193.
- Mizrachi, E.; Myburg, A.A. Systems genetics of wood formation. Curr. Opin. Plant Biol. 2016, 30, 94–100.
- Chiang, M.H.; Greb, T. How to organize bidirectional tissue production? Curr. Opin. Plant Biol. 2019, 51, 15–21.
- Wang, D.; Chen, Y.; Li, W.; Li, Q.; Lu, M.; Zhou, G.; Chai, G. Vascular Cambium: The Source of Wood Formation. Front. Plant Sci. 2021, 12, 700928.
- Lymperopoulos, P.; Msanne, J.; Rabara, R. Phytochrome and Phytohormones: Working in Tandem for Plant Growth and Development. Front. Plant Sci. 2018, 9, 1037.
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859.
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259.
- Nilsson, J.; Karlberg, A.; Antti, H.; Lopez-Vernaza, M.; Mellerowicz, E.; Perrot-Rechenmann, C.; Sandberg, G.; Bhalerao, R.P. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell 2008, 20, 843–855.
- Ibañes, M.; Fàbregas, N.; Chory, J.; Caño-Delgado, A.I. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc. Natl. Acad. Sci. USA 2009, 106, 13630–13635.
- Immanen, J.; Nieminen, K.; Smolander, O.P.; Kojima, M.; Alonso Serra, J.; Koskinen, P.; Zhang, J.; Elo, A.; Mähönen, A.P.; Street, N.; et al. Cytokinin and Auxin Display Distinct but Interconnected Distribution and Signaling Profiles to Stimulate Cambial Activity. Curr. Biol. 2016, 26, 1990–1997.
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574.
- Smetana, O.; Mäkilä, R.; Lyu, M.; Amiryousefi, A.; Sánchez Rodríguez, F.; Wu, M.F.; Solé-Gil, A.; Leal Gavarrón, M.; Siligato, R.; Miyashima, S.; et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 2019, 565, 485–489.
- Fischer, U.; Kucukoglu, M.; Helariutta, Y.; Bhalerao, R.P. The Dynamics of Cambial Stem Cell Activity. Annu. Rev. Plant Biol. 2019, 29, 293–319.
- Xu, C.; Shen, Y.; He, F.; Fu, X.; Yu, H.; Lu, W.; Li, Y.; Li, C.; Fan, D.; Wang, H.C.; et al. Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol. 2019, 222, 752–767.
- Zhu, Y.; Song, D.; Xu, P.; Sun, J.; Li, L. A HD-ZIP III gene, PtrHB4, is required for interfascicular cambium development in Populus. Plant Biotechnol. J. 2018, 16, 808–817.
- Tang, X.; Wang, D.; Liu, Y.; Lu, M.; Zhuang, Y.; Xie, Z.; Wang, C.; Wang, S.; Kong, Y.; Chai, G.; et al. Dual regulation of xylem formation by an auxin-mediated PaC3H17-PaMYB199 module in Populus. New Phytol. 2020, 225, 1545–1561.
- Zheng, S.; He, J.; Lin, Z.; Zhu, Y.; Sun, J.; Li, L. Two MADS-box genes regulate vascular cambium activity and secondary growth by modulating auxin homeostasis in Populus. Plant Commun. 2021, 2, 100134.
- Nieminen, K.; Immanen, J.; Laxell, M.; Kauppinen, L.; Tarkowski, P.; Dolezal, K.; Tähtiharju, S.; Elo, A.; Decourteix, M.; Ljung, K.; et al. Cytokinin signaling regulates cambial development in poplar. Proc. Natl. Acad. Sci. USA 2008, 105, 20032–20037.
- Ohashi-Ito, K.; Saegusa, M.; Iwamoto, K.; Oda, Y.; Katayama, H.; Kojima, M.; Sakakibara, H.; Fukuda, H. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr. Biol. 2014, 8, 2053–2058.
- Smet, W.; Sevilem, I.; de Luis Balaguer, M.A.; Wybouw, B.; Mor, E.; Miyashima, S.; Blob, B.; Roszak, P.; Jacobs, T.B.; Boekschoten, M.; et al. DOF2.1 Controls Cytokinin-Dependent Vascular Cell Proliferation Downstream of TMO5/LHW. Curr. Biol. 2019, 4, 520–529.
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351.
- Kondo, Y.; Ito, T.; Nakagami, H.; Hirakawa, Y.; Saito, M.; Tamaki, T.; Shirasu, K.; Fukuda, H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 2014, 24, 3504.
- Du, J.; Gerttula, S.; Li, Z.; Zhao, S.T.; Liu, Y.L.; Liu, Y.; Lu, M.Z.; Groover, A.T. Brassinosteroid regulation of wood formation in poplar. New Phytol. 2020, 225, 1516–1530.
- Lee, J.; Han, S.; Lee, H.Y.; Jeong, B.; Heo, T.Y.; Hyun, T.K.; Kim, K.; Je, B.I.; Lee, H.; Shim, D.; et al. Brassinosteroids facilitate xylem differentiation and wood formation in tomato. Planta 2019, 249, 1391–1403.
- Lee, J.; Kim, H.; Park, S.G.; Hwang, H.; Yoo, S.I.; Bae, W.; Kim, E.; Kim, J.; Lee, H.Y.; Heo, T.Y.; et al. Brassinosteroid-BZR1/2-WAT1 module determines the high level of auxin signalling in vascular cambium during wood formation. New Phytol. 2021, 230, 1503–1516.
- Love, J.; Björklund, S.; Vahala, J.; Hertzberg, M.; Kangasjärvi, J.; Sundberg, B. Ethylene is an endogenous stimulator of cell division in the cambial meristem of Populus. Proc. Natl. Acad. Sci. USA 2009, 106, 5984–5989.
- Yang, S.; Wang, S.; Li, S.; Du, Q.; Qi, L.; Wang, W.; Chen, J.; Wang, H. Activation of ACS7 in Arabidopsis affects vascular development and demonstrates a link between ethylene synthesis and cambial activity. J. Exp. Bot. 2020, 71, 7160–7170.
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 2000, 18, 784–788.
- Park, E.J.; Kim, H.T.; Choi, Y.I.; Lee, C.; Nguyen, V.P.; Jeon, H.W.; Cho, J.S.; Funada, R.; Pharis, R.P.; Kurepin, L.V.; et al. Overexpression of gibberellin 20-oxidase1 from Pinus densiflora results in enhanced wood formation with gelatinous fiber development in a transgenic hybrid poplar. Tree Physiol. 2015, 35, 1264–1277.
- Jeon, H.W.; Cho, J.S.; Park, E.J.; Han, K.H.; Choi, Y.I.; Ko, J.H. Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 2016, 14, 1161–1170.
- Mauriat, M.; Moritz, T. Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 2009, 58, 989–1003.
- Denis, E.; Kbiri, N.; Mary, V.; Claisse, G.; Conde e Silva, N.; Kreis, M.; Deveaux, Y. WOX 14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 2017, 90, 560–572.
- Ito, Y.; Nakanomyo, I.; Motose, H.; Iwamoto, K.; Sawa, S.; Dohmae, N.; Fukuda, H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 2006, 313, 842–845.
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213.
- Etchells, J.P.; Turner, S.R. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 2010, 137, 767–774.
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 2010, 22, 2618–2629.
- Etchells, J.P.; Mishra, L.S.; Kumar, M.; Campbell, L.; Turner, S.R. Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr Biol. 2015, 25, 1050–1055.
- Kucukoglu, M.; Nilsson, J.; Zheng, B.; Chaabouni, S.; Nilsson, O. WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 2017, 215, 642–657.
- Zhu, Y.; Song, D.; Zhang, R.; Luo, L.; Cao, S.; Huang, C.; Sun, J.; Gui, J.; Li, L. A xylem-produced peptide PtrCLE20 inhibits vascular cambium activity in Populus. Plant Biotechnol J. 2020, 18, 195–206.
- Saito, M.; Kondo, Y.; Fukuda, H. BES1 and BZR1 Redundantly Promote Phloem and Xylem Differentiation. Plant Cell Physiol. 2018, 59, 590–600.
- Han, S.; Cho, H.; Noh, J.; Qi, J.; Jung, H.J.; Nam, H.; Lee, S.; Hwang, D.; Greb, T.; Hwang, I. BIL1-mediated MP phosphorylation integrates PXY and cytokinin signalling in secondary growth. Nat. Plants 2018, 4, 605–614.
- Robischon, M.; Du, J.; Miura, E.; Groover, A. The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 2011, 155, 1214–1225.
- Du, J.; Miura, E.; Robischon, M.; Martinez, C.; Groover, A. The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE 2011, 6, e17458.
- Zhu, Y.; Song, D.; Sun, J.; Wang, X.; Li, L. PtrHB7, a class III HD-Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol. Plant 2013, 6, 1331–1343.
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87.
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860.
- Zhou, J.; Zhong, R.; Ye, Z.H. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS ONE 2014, 9, e105726.
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC Transcription Factors, NST1 and NST3, Are Key Regulators of the Formation of Secondary Walls in Woody Tissues of Arabidopsis. Plant Cell 2007, 19, 270–280.
- Zhong, R.; Richardson, E.A.; Ye, Z.H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611.
- Tan, T.T.; Endo, H.; Sano, R.; Kurata, T.; Yamaguchi, M.; Ohtani, M.; Demura, T. Transcription Factors VND1-VND3 Contribute to Cotyledon Xylem Vessel Formation. Plant Physiol. 2018, 176, 773–789.
- Ohtani, M.; Nishikubo, N.; Xu, B.; Yamaguchi, M.; Mitsuda, N.; Goué, N.; Shi, F.; Ohme-Takagi, M.; Demura, T. A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 2011, 67, 499–512.
- Yang, Y.; Yoo, C.G.; Rottmann, W.; Winkeler, K.A.; Collins, C.M.; Gunter, L.E.; Jawdy, S.S.; Yang, X.; Pu, Y.; Ragauskas, A.J.; et al. PdWND3A, a wood-associated NAC domain-containing protein, affects lignin biosynthesis and composition in Populus. BMC Plant Biol. 2019, 11, 486.
- Takata, N.; Awano, T.; Nakata, M.T.; Sano, Y.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol. 2019, 39, 514–525.
- Yamaguchi, M.; Mitsuda, N.; Ohtani, M.; Ohme-Takagi, M.; Kato, K.; Demura, T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011, 66, 579–590.
- Endo, H.; Yamaguchi, M.; Tamura, T.; Nakano, Y.; Nishikubo, N.; Yoneda, A.; Kato, K.; Kubo, M.; Kajita, S.; Katayama, Y.; et al. Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 2015, 56, 242–254.
- Ohashi-Ito, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 2010, 22, 3461–3473.
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288.
- Ohtani, M.; Demura, T. The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 2019, 56, 82–87.
- Kim, W.C.; Ko, J.H.; Han, K.H. Identification of a cis-acting regulatory motif recognized by MYB46, a master regulator of secondary wall biosynthesis. Plant Mol. Biol. 2012, 78, 489–501.
- Ko, J.H.; Jeon, H.W.; Kim, W.C.; Han, K.H. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 2014, 114, 1099–1107.
- Yang, J.H.; Lee, K.H.; Du, Q.; Yang, S.; Yuan, B.; Qi, L.; Wang, H. A membrane-associated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity. New Phytol. 2020, 226, 59–74.
This entry is offline, you can click here to edit this entry!
