Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The genome sequence of any organism is key to understanding the biology and utility of that organism. Plants have diverse, complex and sometimes very large nuclear genomes, mitochondrial genomes and much smaller and more highly conserved chloroplast genomes. Plant genome sequences underpin our understanding of plant biology and serve as a key platform for the genetic selection and improvement of crop plants to achieve food security.
- DNA sequencing
- plant genome
- long read sequencing
1. Introduction
Advances in the analysis of DNA sequences have been a key driver of enhanced biological understanding and the application of biological knowledge [1]. DNA sequencing in the 20th century was largely based on Sanger sequencing, which limited both the quality (accuracy) and volume of data that could be generated relative to the next generation sequencing that we have today [2]. The introduction and rapid development of next generation sequencing has resulted in an acceleration in the development of plant genome sequencing, especially over the last decade [3]. This technology has evolved rapidly, resulting in continuous major changes to the strategies that are used to sequence and assemble genomes. For example, when only short-read sequences were available, physical mapping was a key strategy. Large fragments of the genomes were cloned in bacterial artificial chromosomes (BACs) [4]. The BACs were then sequenced and the genomes were assembled by covering the genetic maps with BAC tiles [5]. The availability of accurate long read sequencing has made these approaches largely redundant [6]. A review in 2018 [7] reported that 236 angiosperm genome sequences had been reported. Since then, many more genomes have been sequenced and the quality of the genome sequences has increased significantly. The NCBI database (https://www.ncbi.nlm.nih.gov/genome/browse#!/overview/flowering%20plants; accessed 6 June 2022) includes 831 flowering plant genomes, with 373 at the chromosome level. The de novo assembly of long read sequences allows very large contigs to be assembled, sometimes representing a complete plant chromosome [8].
The technology that is now available for plant genome sequencing and assembly make this an increasingly cost-effective strategy for improving our understanding of the biology of all plant species and a key tool for the conservation of plant biodiversity and the use of plants in agriculture and food production. The sequencing of all plant species is a long-term goal that may become key to effectively supporting life on Earth through the improved management of plants in wild populations and their selection and genetic enhancement for use in agriculture and food production.
2. Diversity of Plant Genomes
Plant genomes vary enormously in size, even within closely related groups of plants [9]. The nuclear genomes of flowering plants (angiosperms) vary more than 1000-fold, from less than 100 kb to more than 100 Gb [10]. The genomes of gymnosperms are generally large and complex and represent an even greater challenge for genome sequencing [11]. The large (10 Gb) genome of Ginkgo biloba has recently been reported [12], which provides the first reference genome for gymnosperms. Genomes also vary greatly in terms of their content of repetitive sequences, the level of gene duplication, their ploidy and their heterozygosity, providing a range of challenges and degrees of difficulty within genome sequencing and assembly.
3. Applications of Plant Genome Sequencing
3.1. Model Genomes
The challenge of sequencing plant genomes using early technologies made it necessary to focus on sequencing model genomes that could be used to study related, but more complex, species. The first plant to have a sequenced genome was Arabidopsis thaliana [13], which was chosen because it is a small plant with a rapid generation time and a very small genome, thereby making it an ideal model plant for research use. The first crop plant with a sequenced genome [14] was rice (Oryza sativa), which was chosen because it is a major food crop plant with a relatively small genome. This became a model for cereal and grass genomes. Similarly, Brachypodium distachyon was sequenced [15] as a model grass genome, which is especially relevant for the wheat genome. Recent advances in genome sequencing technology have greatly reduced the need for models as it is now possible to sequence most species easily.
3.2. Crop Plant Genomes
The sequencing of the genomes of crop species has become a key enabling tool for plant improvement. Most major crops now have reference genome sequences [16] and as the technology becomes more powerful and the costs reduce, genomes are also being generated for many other minor crops. This usually involves the production of a reference genome sequence for a species and the re-sequencing of many individuals to define allelic variations within that species. Current efforts recognize that a single reference genome cannot always serve the needs of plant breeders, so pan-genomes that capture the variations in many diverse genomes within the gene pool are being produced as breeding platforms.
3.3. Sequencing Plant Biodiversity
Many diverse plant genomes have now been sequenced with an increasing coverage of the major groups, especially among flowering plants. The coverage of plant orders is high and the genomes from many plant families have now been reported; however, coverage at the genus level is still very low for most plant groups. Systematic efforts to obtain plant genome sequences may take a top-down approach to sequencing a member of each plant family, then each genus and, finally, each species would become available as resources. Ultimately, the re-sequencing of the diversity within each species is of value. A knowledge of the diversity within plant populations is a fundamental tool that can guide the effective conservation of the diversity within species.
3.4. Sequencing Rare and Threatened Species
Targeted efforts are now being made to sequence rare and threatened species of plants as a tool to aid conservation, both in situ [17] and ex situ [18]. This is more urgent among critically endangered species, for which a genome sequence may be all we can retain as the species are lost to extinction. Efforts to sequence biodiversity often focus on rare species as the highest priority.
The critically endangered wild crop relative Macadamia jansenii has been used to compare plant genome sequencing and assembly methods [19]. This has allowed for the comparison of sequencing platforms and bioinformatics tools for genome assembly using a common sample. The generation of a chromosome-level genome sequence for a plant involves the preparation of a DNA sample, the sequencing of that DNA, the assembly of the sequence reads into contigs and, finally, the assembly of the sequence contigs into a chromosome-level assembly (Figure 1).
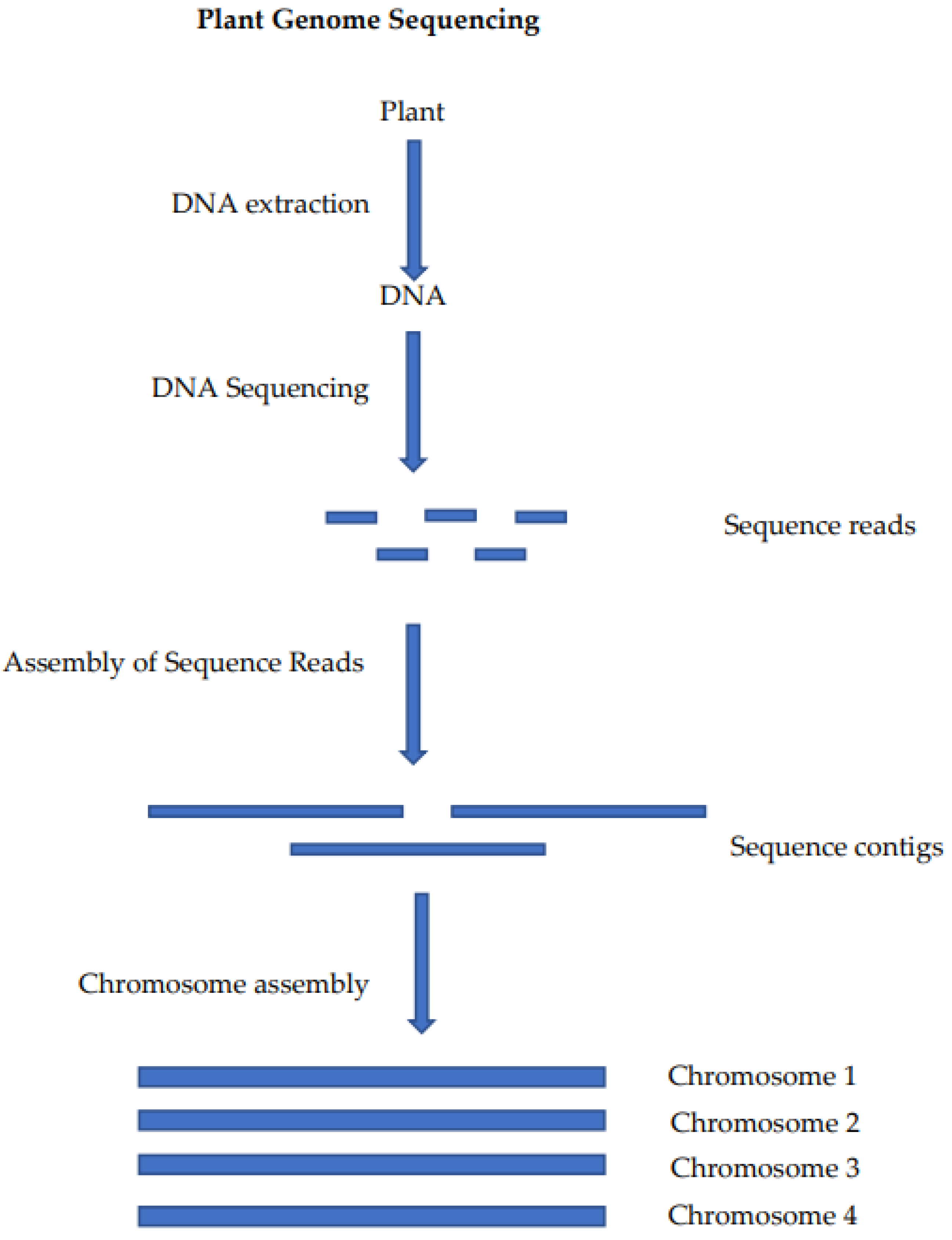
Figure 1. Steps in the sequencing and assembly of a plant genome: DNA extraction is used to produce a DNA sample that is suitable for sequencing, the sequencing of the DNA produces long read sequences, the reads are self-assembled into contigs (often at or near chromosome length) and these contigs are then assembled at the chromosome level using chromatin mapping or genetic mapping.
This entry is adapted from the peer-reviewed paper 10.3390/applbiosci1020008
References
- Henry, R.J. Applied Biosciences: Application of Biological Science and Technology. Appl. Biosci. 2022, 1, 38–39.
- Shendure, J.; Balasubramanian, S.; Church, G.M.; Gilbert, W.; Rogers, J.; Schloss, J.; Waterston, R.H. DNA sequencing at 40: Past, present and future. Nature 2017, 550, 345–353.
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and participation across 20 years of plant genome sequencing. Nat. Plants 2021, 7, 1571–1578.
- Yüksel, B.; Paterson, A.H. Construction and characterization of a peanut HindIII BAC library. Theor. Appl. Genet. 2005, 111, 630–639.
- Garsmeur, O.; Droc, G.; Antonise, R.; Grimwood, J.; Potier, B.; Aitken, K.; Jenkins, J.; Martin, G.; Charron, C.; Hervouet, C.; et al. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 2018, 9, 2638.
- Pucker, B.; Irisarri, I.; de Vries, J.; Xu, B. Plant genome sequence assembly in the era of long reads: Progress, challenges and future directions. Quant. Plant Biol. 2021, 3, 1–14.
- Chen, F.; Dong, W.; Zhang, J.; Guo, X.; Chen, J.; Wang, Z.; Lin, Z.; Tang, H.; Zhang, L. The Sequenced Angiosperm Genomes and Genome Databases. Front. Plant Sci. 2018, 9, 418.
- Sharma, P.A.O.; Alsubaie, B.; Al-Mssallem, I.; Nath, O.; Mitter, N.; Margarido, G.R.A.; Topp, B.; Murigneux, V.; Masouleh, A.K.; Furtado, A.; et al. Improvements in The Sequencing and Assembly of Plant Genomes. Gigabyte 2021.
- Wendel, J.F.; Jackson, S.A.; Meyers, B.C.; Wing, R.A. Evolution of plant genome architecture. Genome Biol. 2016, 17, 37.
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes 2018, 9, 88.
- Uddenberg, D.; Akhter, S.; Ramachandran, P.; Sundström, J.F.; Carlsbecker, A. Sequenced genomes and rapidly emerging technologies pave the way for conifer evolutionary developmental biology. Front. Plant Sci. 2015, 6, 970.
- Liu, H.; Wang, X.; Wang, G.; Cui, P.; Wu, S.; Ai, C.; Hu, N.; Li, A.; He, B.; Shao, X.; et al. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat. Plants 2021, 7, 748–756.
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X.Y.; et al. Analysis of The Genome Sequence of The Flowering Plant Arabidopsis Thaliana. Nature 2000, 408, 796–815.
- Jackson, S.A. Rice: The First Crop Genome. Rice 2016, 9, 14.
- Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; et al. Genome Sequencing and Analysis of the Model Grass Brachypodium Distachyon. Nature 2010, 463, 763–768.
- Kersey, P.J. Plant genome sequences: Past, present, future. Curr. Opin. Plant Biol. 2018, 48, 1–8.
- Wambugu, P.W.; Henry, R.; Browne, L. Supporting in situ conservation of the genetic diversity of crop wild relatives using genomic technologies. Mol. Ecol. 2022, 31, 2207–2222.
- Wambugu, P.W.; Ndjiondjop, M.-N.; Henry, R.J. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief. Funct. Genom. 2018, 17, 198–206.
- Murigneux, V.; Rai, S.K.; Furtado, A.; Bruxner, T.J.C.; Tian, W.; Harliwong, I.; Wei, H.; Yang, B.; Ye, Q.; Anderson, E.; et al. Comparison of long-read methods for sequencing and assembly of a plant genome. GigaScience 2020, 9, giaa146.
This entry is offline, you can click here to edit this entry!
