Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Since the outbreak of coronavirus disease-19 (COVID-19), cold-chain food contamination caused by the pathogenic severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has attracted huge concern. Cold-chain foods provide a congenial environment for SARS-CoV-2 survival, which presents a potential risk for public health. Strengthening the SARS-CoV-2 supervision of cold-chain foods has become the top priority in many countries
- SARS-CoV-2
- cold-chain foods
- RT-LAMP
1. Potential Cold-Chain Food Quarantine Techniques
Unlike traditional food testing methods, the detection of SARS-CoV-2 on cold-chain foods puts forward higher requirements regarding sampling method, detecting conditions, testing periodic time, anti-interference capability, and portability of equipment [1]. Here, according to the different pathogen markers, e.g., nucleic acid, antigen, antibody, and cytokine storm assay, researchers list two categories of COVID-19 testing methods, as well as the advance of SARS-CoV-2 test methods (Table 1). Whether the existing detection methods can be applied to test suspicious cold-chain foods will be dialectically discussed.
Table 1. The advance of SARS-CoV-2 test techniques.
| Methods | Category | Subcategory | LOD | Specificity | Sensitivity | Cost | Time | Description | References |
|---|---|---|---|---|---|---|---|---|---|
| Nucleic acid test | RT-PCR | 1–10 copies | 97.06–99.69% | 91.06–99.96% | USD 25–200 | 4-6 h | The gold standard for SARS-CoV-2 diagnosis is suitable for the large-scale test but needs specialized laboratory equipment and trained technicians | [2][3] | |
| Whole-genome sequencing | ND | ND | 98.33–99.83% | USD 2000 | 48-72 h | The first complete genomic sequences of SARS-CoV-2 were obtained through metatranscriptomics approaches | [4][5] | ||
| Isothermal amplification technology | Transcriptional colorimetric loop-mediated isothermal amplification | 100 copies/μL | 100% | 85% | ND | 21 h | Effectively reduce the false positive rate and improve the detection efficiency | [6] | |
| Proofreading enzyme-mediated isothermal amplification | 100 copies | Effectively distinguish SARS-CoV-2 from SARS-CoV | Effectively detect as few as 100 copies of gene N RNA in 1 h | ND | 50 min | Show similar analytical performance with the conventional RT-PCR | [7] | ||
| Emulsion loop-mediated isothermal amplification | 10, 103, and 105 copies/μL | ND | ND | ND | 5–10 min | Limit of detection of 1 copy per microliter sample and portable device using a miniature spectrometer or a smartphone | [8] | ||
| Recombinase polymerase amplification (RPA) |
Combined RPA with rkDNA-graphene oxide probing system | 6.0 aM | ND | ND | ND | 1.6 h | Exhibit high selectivity and sensitivity for the diagnosis of COVID-19 | [9] | |
| Recombinase polymerase amplification | 7.659 copies/μL | 100% | 98% | USD 4.3 | 5–20 min | High specificity | [10][11] | ||
| Isothermal RPA-lateral flow detection | 0.25–2.5 copies/μL | 100% | 94% | ND | 5 min | The detection limit of RPA-LF for SARS-CoV-2 was 35.4 nucleocapsid (N) gene copies/L; the sensitivity was similar to that of qualitative real-time PCR | [12] | ||
| Hybrid capture immunofluorescence assay | Hybrid capture immunofluorescence assay | 500 copies per mL | 99% | 100% | ND | 45 min | The detection sensitivity is consistent with similar products on the market; however, this technique can only give qualitative results | [13] | |
| Entropy-driven amplified electrochemiluminescence | 2.67 fM | ND | ND | ND | 10–20 h | High selectivity and stability | [14] | ||
| CRISPR-based test | Cas12a | 10 copies per μL reaction | 100% | 95% | USD 6 | 40–60 min | Enables rapid, ultrasensitive (few copies), and highly specific nucleic acid detections | [15] | |
| Cas13a | 10–100 copies per μL | 100% | 96% | USD 3.5 | 40–57 min | Rapid, sensitive, and with low instrument requirement | [16] | ||
| Pyrococcus furiosus Argonaute coupled with modified ligase chain reaction | 10 aM | ND | ND | Cheaper than CRISPR | ~70 min | High sensitivity, high specificity, and multiplexing detection; without the use of RNA as guidance | [17] | ||
| Immunological test | Antigen immunological test | Quantum dot immunochromatographic assay | 4.9 pg/mL | 100% | 75 pg/mL | USD 1.5 | 3 min | One single test that can cover hs-CRP and routine-range CRP with a detection range from 1 to 200 μg mL−1 | [18] |
| QuickNavi™-COVID-19 Ag immunochromatographic test | ND | 100% | 86.7% | Cheaper than nucleic acid amplification tests | 5 min | The overall sensitivity was 86.7%, and the positive detection rate in patients with CT < 30 was comparable to that of RT-PCR | [19] | ||
| Magnetic graphene quantum dots | 248 Particles mL−1 | Related to SARS-CoV-2 antigen protein | No response to MERS-CoV | USD 1.25 | 2 min | Sensitive detection without sample pretreatment in one step with a LOD of 248 Particles mL−1 | [20] | ||
| Binax-CoV2 | 1.6 × 104–4.3 × 104 viral RNA copies | 99.9% | 93.3% | USD 5 | 15 min | The sensitivity of Binax-CoV2 was 93.3% and the specificity was 99.9% | [21] | ||
| SERS biosensor | 80 copies mL−1 | Related to the sensing environment | Suffers from non-specific binding | More expensive than ELISA | 5 min | The low detection limit (LOD) can be reduced to 80 parts mL−1 | [22] | ||
| Interdigitated microelectrode chip | 2.29 × 10−6 ng/mL | 4.27 × 10−4 ng/mL | 234:1 | USD 1 | 20 s | The linear range is 10−5–10−1 ng/mL; the strategy is real-time, sensitive, selective, and large-scale in cold-chain food quarantine | [23] | ||
| Serum antibody immunological test | Split luciferase antibody biosensors | ND | > 99% | > 98% | ∼15 ¢ | 5 min | The sensitivity to detect anti-S protein antibodies was 89% and anti-N protein antibodies were 98%, and the specificity of both was more than 99% | [24] | |
| Colloidal gold immunochromatography assay | 20.00 IU/mL | 96.2% | 71.1% | ND | 10–15 min | The IgM/IgG test assay demonstrated high sensitivity of 71.1% and specificity of 96.2% in 150 suspect COVID-19 cases | [25] | ||
| Chemiluminescence immunoassay | 0.5–1.5 AU/mL | 97.5% | 78.65% | ND | 1 h | The antibody detection rate has high sensitivity, high precision, quantitative detection, and easy automation | [26] | ||
| Upconverting phosphor immunochromatography assay | ND | 99.75% | 89.15% | ND | 10 min | High sensitivity, no interference from the background, and good stability | [27] | ||
| Surface plasmon resonance biosensors (SPRS) | 0.22 pM | ND | ND | ND | ND | The SPR biosensor is feasible in the concentration range of 2 to 1000 ng/mL | [28] | ||
| DNA-assisted nanopore sensing | 50 ng/mL (IgM) 10 ng/mL(IgG) | ND | ND | USD 8 | ND | High sensitivity and specificity compared to laboratory techniques | [29] | ||
| Colorimetric-fluorescent dual-mode lateral flow immunoassay biosensor | 10 ng/mL(IgM) 5 ng/mL(IgG) | 100% | ND | ND | ND | The combined detection sensitivity and specificity of this assay for IgM/IgG is 100%, and it has great potential for rapid and accurate detection | [29] | ||
| The lateral flow immunoassay method | ND | 90.63% | 88.66% | ND | 15 min | The limits of detection for IgM and IgG were 10 ng/mL and 5 ng/mL, respectively | [30] | ||
| Luciferase immunosorbent assay (LISA) | 0.4–75 pg / μl | 100% | 71% | ND | ~60 min | LISA had a sensitivity of 71% in COVID-19 patients and a specificity of 100% in healthy blood donors in the second week after onset | [31] | ||
| Enzyme-linked immunosorbent assays | 0.095 (IgM) 0.083 (IgG) | ND | 98% | ND | 80–120 min | High sensitivity and specificity | [32] | ||
| Enzyme-linked immunosorbent assays | ND | 88.2–99.2% (IgM) 75.6–98.3% (IgG) | 78.2% (IgG) 96.6%(IgM) | ND | 1.5 h | ELISA was used to detect IgG antibodies in confirmed patients with COVID-19, and the sensitivity to detect IgM antibodies was low | [33] | ||
| Enzyme-linked immunosorbent assays (ELISA) | ND | 93–100% | 65–85% | ND | 4 h | The detection precision is similar to ELISA, but the detection range is wider and the sensitivity is higher | [34] |
Note that ND is not defined in the literature. LOD: limit of detection. RT-PCR(Reverse transcription-polymerase chain reaction). CRISPR(Clustered Regularly Interspaced Short Palindromic Repeats), SERS(Surface-enhanced raman scattering). aM: 10−18 mol/mL. fM: 10−15 mol/mL. IU: international unit. AU: arbitrary unit.
2. Nucleic Acid Test
SARS-CoV-2 is a positive-sense RNA virus, and the feature gene can be used as target analytes [35]. Nucleic acid detection technology is the most direct and essential pathogenic evidence for food contamination (Figure 1A). It has the advantages of early diagnosis, high sensitivity, and good specificity, and is the gold standard for SARS-CoV-2 detection (e.g., RT-PCR) [36].
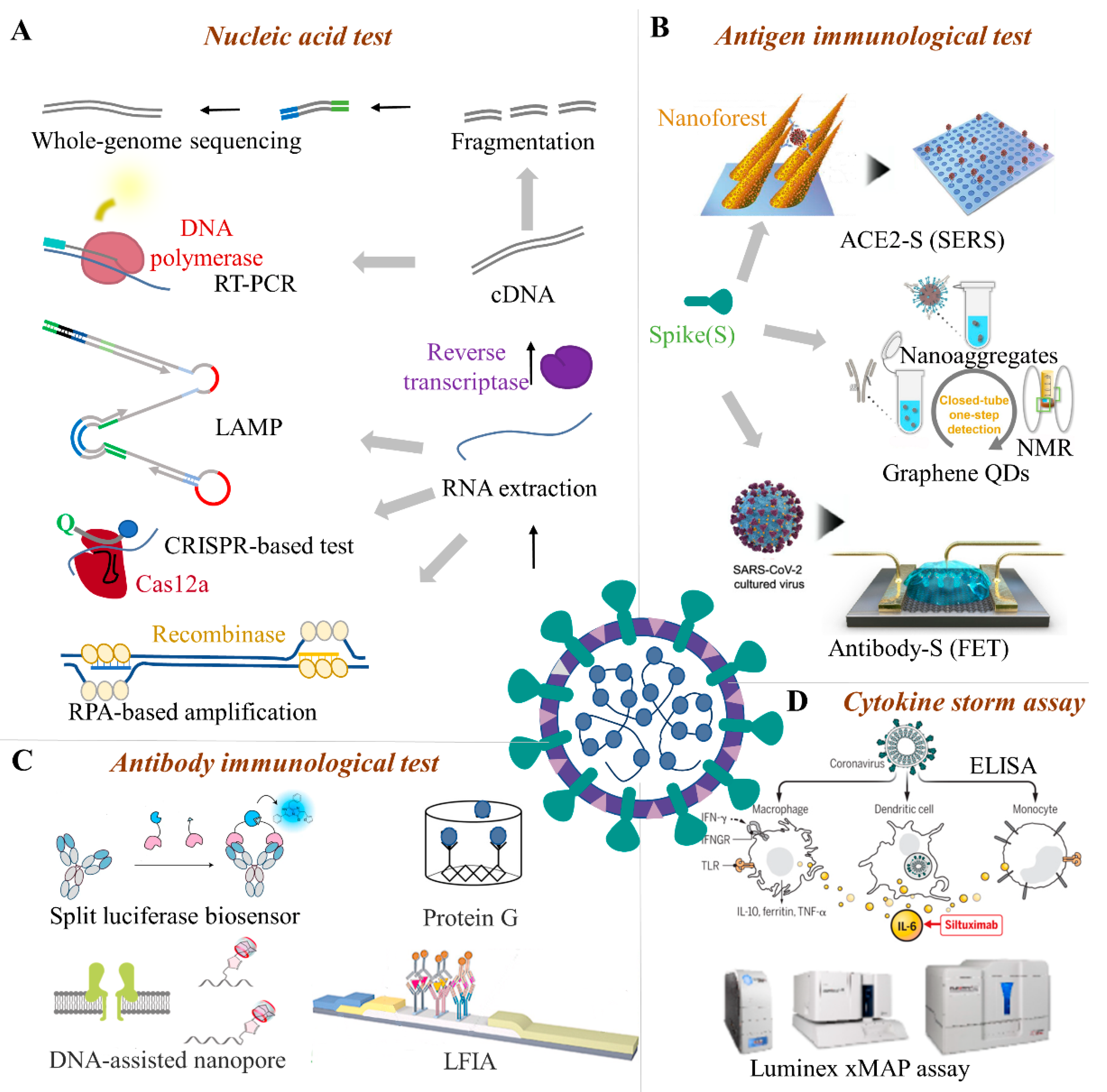
Figure 1. The advanced quarantine methods for SARS-CoV-2. For SARS-CoV-2 quarantine, many test techniques including nucleic acid and immunological methods are available. Nucleic acid tests include whole-genome sequencing and specific gene detection (A). Immunological tests include antigen tests (B), antibody immunological tests (C), and cytokine storm diagnoses (D). RT-PCR(Reverse transcription-polymerase chain reaction), LAMP (Loop-mediated isothermal amplification), CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), LFIA (Lateral-flow immunochromatographic assay), FET (Field-effect transistor), ELISA (Enzyme linked immunosorbent assay), xMAP (Multi-analyte profiling), SERS(Surface-enhanced raman scattering).
2.1. PCR-Based Techniques
Among the nucleic acid detection methods, RT-PCR can effectively amplify trace viral genes in nucleic acid mixtures and has the characteristics of fast detection speed, high sensitivity, and strong specificity [37]. RT-PCR uses sequence-specific primers to identify tiny RNA targets. The recognized RNA is then transcribed by reverse transcriptase to cDNA, which is then used as a template for DNA replication through PCR. However, the measurement is an enzyme-dependent multi-step technique, and the operation is complicated. The turnaround time takes a few hours which cannot meet the requirements of rapid testing of cold-chain food [38]. The test facilities and instruments are not portable, and the collected samples need to be transported to the laboratory for testing. The samples may produce false-negative results due to improper collection or processing. Improper operation or insufficient laboratory conditions may cause false positives due to aerosol contamination [39][40][41]. One-step nested RT-PCR is a flexible and easy method to test SARS-CoV-2. If the coronavirus mutates in one key amplified nucleotide, at least one pair can still be amplified [42]. The detection cost is lower than RT-PCR, but nested PCR is not feasible as a detection method for cold-chain food, because it is time-consuming and has a high risk of cross-contamination. In contrast, repetitive digital PCR is less interfered with by background wild DNA molecules, so it can reduce the impact of non-target DNA during cold-chain detection [43]. Digital PCR has obvious advantages when the viral load of cold-chain food samples is low or the sample nucleic acid is degraded. However, the cost of digital PCR detection is relatively high and the instrument is not portable [44]. It also involves the usage and storage of enzyme reagents, which will increase the cost and technological hurdles of detection. The digital PCR operation process has the disadvantage of being easily contaminated. To avoid false-positive results, it is necessary to establish strict internal quality control specifications for the laboratory and strictly regulate the testing operation process [45][46].
2.2. RT-LAMP
Reverse transcription loop-mediated isothermal amplification (RT-LAMP) is a nucleic acid amplification assay, which is characterized by multiple specific primers for the target gene at a constant temperature of 60–65 °C under the DNA polymerase. About 109-1010 times of nucleic acid amplification can be achieved in 15–60 min. RT-LAMP has the characteristics of simple operation, strong specificity (2 to 5 orders of magnitude higher than traditional PCR methods), and easy product detection [47]. The RT-LAMP results can be judged by visually observing the generation of white turbidity or green fluorescence. It is simple, quick, and does not require gel electrophoresis like PCR. As a point-of-care testing (POCT)-type nucleic acid detection method, LAMP requires less professional equipment, such as a thermal cycler, and the price of the instrument is lower than qRT-PCR, which can well meet the real-time requirements of SARS-CoV-2 detection on cold-chain food [48]. The detection sensitivity of RT-LAMP can reach 10 copies, and it has high specificity [49]. RT-LAMP is highly suitable for detecting > 60 copies/10 μL in sample. However, the test involves the use of enzymes (e.g., recombinase) that the storage of enzyme reagent needs, resulting in extra cost. When the test is performed for a long time, non-specific amplification may produce false-positive results if cold-chain food sampling < 10 copies/10 μL [50]. Based on RT-LAMP, a portable and scalable laser-engraved microwell array chip for multiplex amplification of viral RNA samples has been developed, which is a promising device for SARS-CoV-2 detection on cold-chain food.
2.3. CRISPR-Based System
CRISPR-Cas is a nonspecific RNA system that can be activated by the amplified product RNA, cleavages the reporter RNA, and releases a fluorescent dye from the quencher. The CRISPR-based system has attracted growing enthusiasm due to its pathogen diagnosis ability [51]. It can realize the on-site test of SARS-CoV-2 on cold-chain food using simple equipment. The test time varies from 40 to 70 min when excluding the time for RNA extraction [52][53]. Combined with RT-LAMP technology, the CRISPR-Cas system can achieve an LoD of 10 copies/μL. Most CRISPR-based SARS-CoV-2 detection methods use the Cas12 enzyme to specifically recognize the virus sequence [54]. In addition, the all-in-one dual CRISPR-Cas12a analysis system does not need a pre-amplification step and it improves the sensitivity of the assay by using double CRISPR RNA. It can detect 1.2 DNA targets and 4.6 RNA targets in 40 min. The system can be developed as a one-step test platform without the need for cDNA preparation which has the potential for SARS-CoV-2 detection on cold-chain food [55].
2.4. Microfluidic Biochip
Microfluidic chips integrate various small-scale laboratory functions on a single chip to complete the steps in traditional laboratories [56]. It uses a small number of reagents and samples to obtain accurate test results in a short time, and is especially suitable for the rapid detection of SARS-CoV-2 on cold-chain food. Recently, paper-based microfluidics, centrifugal chips, wearable microfluidic devices, and digital nucleic acid detection chips have been proposed for pathogen testing and disease screening [57]. For instance, the IDNOW® instrument proposed by Abbott™ in the United States can detect a positive sample. The product has received an emergency use authorization (EUA) from the U.S. Food and Drug Administration (FDA). The instrument weighs only 3 kg and is portable and suitable for POCT [58][59].
2.5. Whole-Genome Sequencing
Whole-genome sequencing (WGS) is an effective tool to comprehensive understand SARS-CoV-2. The assay belongs to high-throughput sequencing, or next-generation sequencing, which is a culture-free, unbiased, direct extraction of DNA or RNA from clinical samples [3][4]. However, the operation steps of WGS are relatively complex and the operation technology requirements are high. The RNA can be extracted using the kit and whole-genome sequencing performed on an instrument (e.g., Illumina iSeq 100). Peculiarly, metagenomics is a high-sensitivity pan-pathogen assay for the discovery of novel pathogens and infectious disease diagnosis, which is applied in the simultaneous and rapid detection of SARS-CoV-2 [60]. Because the whole gene sequencing requires a professional operator, complex sample pretreatment, and long-term periods, other methods are usually combined to generate test reports.
3. Immunological Methods
SARS-CoV-2 has a wide mammalian host range, including minks, cows, white-tailed deer, dogs, domestic cats, swine, lions, etc. [61][62][63][64]. Some of these animals may serve as viral carriers once they are made into food-related products [65]. Immunological tests can directly detect the antigen biomarker of SARS-CoV-2 and can be used to determine whether food (e.g., animal products) is infected or contaminated by the virus. The immunological methods that mainly include antigen tests, serology tests, and cytokine storm diagnoses can be used for SARS-CoV-2 detection for these animal foods. For plant foods, that cannot undergo an immune response to produce antibodies, serum antibody immunological testing and cytokine storm diagnosis cannot be applied to the detection of cold-chain food unless the plant food is contaminated with the body fluids of an infected person [66]. In the following sections, researchers will describe each of the above methods in detail.
3.1. Antigen Immunological Test
Antigen tests are the main immunological method for food quarantine. The structural proteins, such as the spike glycoprotein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) of SARS-CoV-2 are the primary antigens used for the immunological test. Among the antigen immunological test methods (Figure 1B), ELISA is a highly sensitive immunological experimental technique that combines an antigen, antibody-specific reaction, and high-efficiency enzyme catalysis on the substrate. This detection method has high sensitivity and low difficulty in carrier standardization, but the detection steps are more cumbersome and easier to contaminate [67]. ELISA kits can test multiple samples in a single run; however, they lack point-of-care applicability and the non-specific binding of antibodies or antigens to the plate may lead to false positive results [68]. In addition to ELISA, the detection of trace S-protein (S1 subunit) for real-time SARS-CoV-2 detection is currently a known method that can be well applied to cold-chain foods. The S-protein particles can be attracted to the surface of the sensor and captured by the antibody within 20 s, which meets the real-time detection requirements on-site. The linear range is wide and covers the possible range of the concentration of S-protein on the food surface. The developed ultra-low LOD strategy has shown great advantages in the detection of virus markers with low concentrations of cold-chain foods [23]. The single sensor device can act as a disposable chip and its cost is estimated to be 1 US dollar. The operations of the device are relatively simple and can be operated by non-technical personnel [69]. Thus, the S-protein detection platform meets the requirements of rapid response, lower detection limit, high specificity, friendly operation, and low cost, which provides a promising solution for SARS-CoV-2 detection on cold-chain food [70].
Besides, the biosensor equipped with a chemical or biological receptor (e.g., antibody) can specifically interact with the target analyte showing a quantitative signal of the recognition process [71]. Compared with traditional laboratory methods, biosensors can provide a cheap, sensitive, rapid, miniaturized, and portable platform for SARS-CoV-2 detection which is promising for the on-site detection of cold-chain food contamination. Several studies have proved biosensor technology is convenient in SARS-CoV-2 S protein detection based on a bioelectrical identification assay [72]. The biosensor can detect S protein within 3 min with a LOD of about 1 fg/mL, and no cross-reaction with SARS-CoV-2 nucleocapsid protein was found. The portable readout system of the ready-made biosensor platform can be controlled by a smartphone or a tablet computer [73]. The high sensitivity, rapidness, and simplicity of biosensors make it a great advantage in the detection of viruses on cold-chain food. It can be easily controlled and has strong practical applicability to test contaminations on cold-chain food in factories or customs. However, some biosensors involve the use of enzyme reagents, and the biosensor technology is still in the development stage [74].
3.2. Serum Antibody Immunological Test
Once the body is infected, the living organisms will produce specific antibodies, such as anti-SARS-CoV-2 IgM and IgG. Though animal food is frozen or transported in the cold-chain, the antibodies from an earlier infection can be preserved. The safety risk of infected COVID-19 is relatively low by eating cold-chain animal food and has not been studied to date. However, these serum antibodies can be used as the immunological target, especially in cold-chain animal food with a low viral load. Compared with nucleic acid testing, blood samples for antibody serology testing are easier to obtain, which greatly reduces the risk of infection of medical staff during specimen collection and testing, and makes it easier for primary laboratories to carry out screening work. For instance (Figure 1C), luciferase immunosorbent assay (LISA) is an easily and rapidly developed semi-quantitative method and is appropriate for detecting specific antibodies from cold-chain animal food [31]. Compared with enzyme-linked immunosorbent assays, DNA-assisted nanopore sensing assay can reliably quantify SARS-CoV-2 antibodies with high accuracy, wide dynamic range, and the potential for automated detection on cold-chain food [29]. Though modified with probe DNA to label IgG or IgM antibodies, the nanopore sensor can quantify the probe DNAs when thermal dehybridization of gold nanoparticles (AuNPs) probe DNAs was performed. In addition, surface plasmon resonance (SPR) biosensors assay adopts the optical detection method. Indirect aggregation can be used for virus detection by modifying targeted molecules on the virus surface. If you do not consider the portability of the SPR device, it may be the most promising technique for cold-chain food quarantine [28].
This entry is adapted from the peer-reviewed paper 10.3390/foods11111540
References
- Chi, Y.; Zheng, S.; Liu, C.; Wang, Q. Transmission of SARS-CoV-2 on cold-chain food overpacks: A new challenge. J. Glob. Health 2021, 11, 03071.
- Böger, B.; Fachi, M.M.; Vilhena, R.O.; Cobre, A.F.; Tonin, F.S.; Pontarolo, R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control 2021, 49, 21–29.
- Chaimayo, C.; Kaewnaphan, B.; Tanlieng, N.; Athipanyasilp, N.; Sirijatuphat, R.; Chayakulkeeree, M.; Angkasekwinai, N.; Sutthent, R.; Puangpunngam, N.; Tharmviboonsri, T.; et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol. J. 2022, 17, 177.
- Greninger, A.L.; Zerr, D.M.; Qin, X.; Adler, A.L.; Sampoleo, R.; Kuypers, J.M.; Englund, J.A.; Jerome, K.R. Rapid Metagenomic Next-Generation Sequencing during an Investigation of Hospital-Acquired Human Parainfluenza Virus 3 Infections. J. Clin. Microbiol. 2017, 55, 177–182.
- Chiara, M.; D’Erchia, A.M.; Gissi, C.; Manzari, C.; Parisi, A.; Resta, N.; Zambelli, F.; Picardi, E.; Pavesi, G.; Horner, D.S.; et al. Next generation sequencing of SARS-CoV-2 genomes: Challenges, applications and opportunities. Brief. Bioinform. 2021, 22, 616–630.
- Ji, C.; Xue, S.; Yu, M.; Liu, J.; Zhang, Q.; Zuo, F.; Zheng, Q.; Zhao, L.; Zhang, H.; Cao, J.; et al. Rapid Detection of SARS-CoV-2 Virus Using Dual Reverse Transcriptional Colorimetric Loop-Mediated Isothermal Amplification. ACS Omega 2021, 6, 8837–8849.
- Ding, S.; Chen, G.; Wei, Y.; Dong, J.; Du, F.; Cui, X.; Huang, X.; Tang, Z. Sequence-specific and multiplex detection of COVID-19 virus (SARS-CoV-2) using proofreading enzyme-mediated probe cleavage coupled with isothermal amplification. Biosens. Bioelectron. 2021, 178, 113041.
- Day, A.S.; Ulep, T.H.; Safavinia, B.; Hertenstein, T.; Budiman, E.; Dieckhaus, L.; Yoon, J.Y. Emulsion-based isothermal nucleic acid amplification for rapid SARS-CoV-2 detection via angle-dependent light scatter analysis. Biosens. Bioelectron. 2021, 179, 113099.
- Choi, M.H.; Lee, J.; Seo, Y.J. Combined recombinase polymerase amplification/rkDNA-graphene oxide probing system for detection of SARS-CoV-2. Anal. Chim. Acta 2021, 1158, 338390.
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–958.
- Cherkaoui, D.; Huang, D.; Miller, B.S.; Turbé, V.; McKendry, R.A. Harnessing recombinase polymerase amplification for rapid multi-gene detection of SARS-CoV-2 in resource-limited settings. Biosens. Bioelectron. 2021, 189, 113328.
- Shelite, T.R.; Uscanga-Palomeque, A.C.; Castellanos, A.; Melby, P.C.; Travi, B.L. Isothermal recombinase polymerase amplification-lateral flow detection of SARS-CoV-2, the etiological agent of COVID-19. J. Virol. Methods 2021, 296, 114227.
- Wang, D.; He, S.; Wang, X.; Yan, Y.; Liu, J.; Wu, S.; Liu, S.; Lei, Y.; Chen, M.; Li, L.; et al. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1150–1158.
- Fan, Z.; Yao, B.; Ding, Y.; Zhao, J.; Xie, M.; Zhang, K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021, 178, 113015.
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783.
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705.
- Wang, L.; He, R.; Lv, B.; Yu, X.; Liu, Y.; Yang, J.; Li, W.; Wang, Y.; Zhang, H.; Yan, G.; et al. Pyrococcus furiosus Argonaute coupled with modified ligase chain reaction for detection of SARS-CoV-2 and HPV. Talanta 2021, 227, 122154.
- Zhang, Q.; Li, J.; Li, Y.; Tan, G.; Sun, M.; Shan, Y.; Zhang, Y.; Wang, X.; Song, K.; Shi, R.; et al. SARS-CoV-2 detection using quantum dot fluorescence immunochromatography combined with isothermal amplification and CRISPR/Cas13a. Biosens. Bioelectron. 2022, 202, 113978.
- Takeuchi, Y.; Akashi, Y.; Kato, D.; Kuwahara, M.; Muramatsu, S.; Ueda, A.; Notake, S.; Nakamura, K.; Ishikawa, H.; Suzuki, H. The evaluation of a newly developed antigen test (QuickNavi™-COVID19 Ag) for SARS-CoV-2: A prospective observational study in Japan. J. Infect. Chemother. 2021, 27, 890–894.
- Li, Y.; Ma, P.; Tao, Q.; Krause, H.J.; Yang, S.; Ding, G.; Dong, H.; Xie, X. Magnetic graphene quantum dots facilitate closed-tube one-step detection of SARS-CoV-2 with ultra-low field NMR relaxometry. Sens. Actuators B Chem. 2021, 337, 129786.
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020, 14, 5135–5142.
- Yadav, S.; Sadique, M.A.; Ranjan, P.; Kumar, N.; Singhal, A.; Srivastava, A.K.; Khan, R. SERS based lateral flow immunoassay for point-of-care detection of SARS-CoV-2 in clinical samples. ACS Appl. Bio Mater. 2021, 4, 2974–2995.
- Zhang, J.; Fang, X.; Mao, Y.; Qi, H.; Wu, J.; Liu, X.; You, F.; Zhao, W.; Chen, Y.; Zheng, L. Real-time, selective, and low-cost detection of trace level SARS-CoV-2 spike-protein for cold-chain food quarantine. Npj Sci. Food 2021, 5, 12.
- Elledge, S.K.; Zhou, X.X.; Byrnes, J.R.; Martinko, A.J.; Lui, I.; Pance, K.; Lim, S.A.; Glasgow, J.E.; Glasgow, A.A.; Turcios, K.; et al. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 2020, 39, 928–935.
- Wang, Q.; Du, Q.; Guo, B.; Mu, D.; Lu, X.; Ma, Q.; Guo, Y.; Fang, L.; Zhang, B.; Zhang, G. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. J. Clin. Microbiol. 2020, 58, e00375–e00420.
- Padoan, A.; Cosma, C.; Sciacovelli, L.; Faggian, D.; Plebani, M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin. Chem. Lab. Med. 2020, 58, 1081–1088.
- Niedbala, R.S.; Feindt, H.; Kardos, K.; Vail, T.; Burton, J.; Bielska, B.; Li, S.; Milunic, D.; Bourdelle, P.; Vallejo, R. Detection of analytes by immunoassay using up-converting phosphor technology. Anal. Biochem. 2001, 293, 22–30.
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 2020, 14, 5268–5277.
- Zhang, Z.; Wang, X.; Wei, X.; Zheng, S.W.; Lenhart, B.J.; Xu, P.; Li, J.; Pan, J.; Albrecht, H.; Liu, C. Multiplex quantitative detection of SARS-CoV-2 specific IgG and IgM antibodies based on DNA-assisted nanopore sensing. Biosens. Bioelectron. 2021, 181, 113134.
- Bayin, Q.; Huang, L.; Ren, C.; Fu, Y.; Ma, X.; Guo, J. Anti-SARS-CoV-2 IgG and IgM detection with a GMR based LFIA system. Talanta 2021, 227, 122207.
- Yao, Z.; Drecun, L.; Aboualizadeh, F.; Kim, S.J.; Li, Z.; Wood, H.; Valcourt, E.J.; Manguiat, K.; Plenderleith, S.; Yip, L.; et al. A homogeneous split-luciferase assay for rapid and sensitive detection of anti-SARS CoV-2 antibodies. Nat. Commun. 2021, 12, 1086.
- Bundschuh, C.; Egger, M.; Wiesinger, K.; Gabriel, C.; Clodi, M.; Mueller, T.; Dieplinger, B. Evaluation of the EDI enzyme linked immunosorbent assays for the detection of SARS-CoV-2 IgM and IgG antibodies in human plasma. Clin. Chim. Acta 2020, 509, 79–82.
- Al-Jighefee, H.T.; Yassine, H.M.; Nasrallah, G.K. Evaluation of antibody response in symptomatic and asymptomatic COVID-19 patients and diagnostic assessment of new IgM/IgG ELISA kits. Pathogens 2021, 10, 161.
- Skalnikova, H.K.; Kepkova, K.V.; Vodicka, P. Luminex xMAP Assay to Quantify Cytokines in Cancer Patient Serum. In Immune Mediators in Cancer; Springer: Berlin/Heidelberg, Germany, 2020.
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269.
- Rahbari, R.; Moradi, N.; Abdi, M. rRT-PCR for SARS-CoV-2: Analytical considerations. Clin. Chim. Acta 2021, 516, 1–7.
- Torretta, S.; Zuccotti, G.; Cristofaro, V.; Ettori, J.; Solimeno, L.; Battilocchi, L.; D’Onghia, A.; Bonsembiante, A.; Pignataro, L.; Marchisio, P.; et al. Diagnosis of SARS-CoV-2 by RT-PCR Using Different Sample Sources: Review of the Literature. Ear Nose Throat J. 2021, 100, 131s–138s.
- Sule, W.F.; Oluwayelu, D.O. Real-time RT-PCR for COVID-19 diagnosis: Challenges and prospects. Pan Afr. Med. J. 2020, 35, 121.
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454.
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M.; et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test to Diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438–e01520.
- Yelin, I.; Aharony, N.; Tamar, E.S.; Argoetti, A.; Messer, E.; Berenbaum, D.; Shafran, E.; Kuzli, A.; Gandali, N.; Shkedi, O.; et al. Evaluation of COVID-19 RT-qPCR Test in Multi sample Pools. Clin. Infect. Dis. 2020, 71, 2073–2078.
- Meza-Robles, C.; Barajas-Saucedo, C.E.; Tiburcio-Jimenez, D.; Mokay-Ramírez, K.A.; Melnikov, V.; Rodriguez-Sanchez, I.P.; Martinez-Fierro, M.L.; Garza-Veloz, I.; Zaizar-Fregoso, S.A.; Guzman-Esquivel, J.; et al. One-step nested RT-PCR for COVID-19 detection: A flexible, locally developed test for SARS-CoV2 nucleic acid detection. J. Infect. Dev. Ctries. 2020, 14, 679–684.
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005.
- Anderson, E.M.; Maldarelli, F. Quantification of HIV DNA Using Droplet Digital PCR Techniques. Curr. Protoc. Microbiol. 2018, 51, e62.
- Yin, J.; Hu, J.; Sun, J.; Wang, B.; Mu, Y. A fast nucleic acid extraction system for point-of-care and integration of digital PCR. Analyst 2019, 144, 7032–7040.
- Carraturo, F.; Del Giudice, C.; Morelli, M.; Cerullo, V.; Libralato, G.; Galdiero, E.; Guida, M. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020, 265, 115010.
- Rödel, J.; Egerer, R.; Suleyman, A.; Sommer-Schmid, B.; Baier, M.; Henke, A.; Edel, B.; Löffler, B. Use of the variplex™ SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J. Clin. Virol. 2020, 132, 104616.
- Schermer, B.; Fabretti, F.; Damagnez, M.; Di Cristanziano, V.; Heger, E.; Arjune, S.; Tanner, N.A.; Imhof, T.; Koch, M.; Ladha, A.; et al. Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay. PLoS ONE 2020, 15, e0238612.
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M.; et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961.
- Rabe, B.A.; Cepko, C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. USA 2020, 117, 24450–24458.
- Xiang, X.; Qian, K.; Zhang, Z.; Lin, F.; Xie, Y.; Liu, Y.; Yang, Z. CRISPR-cas systems based molecular diagnostic tool for infectious diseases and emerging 2019 novel coronavirus (COVID-19) pneumonia. J. Drug Target. 2020, 28, 727–731.
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874.
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129.
- Palaz, F.; Kalkan, A.K.; Tozluyurt, A.; Ozsoz, M. CRISPR-based tools: Alternative methods for the diagnosis of COVID-19. Clin. Biochem. 2021, 89, 1–13.
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711.
- Lepej, S.Z.; Poljak, M. Portable molecular diagnostic instruments in microbiology: Current status. Clin. Microbiol. Infect. 2020, 26, 411–420.
- Batule, B.S.; Seok, Y.; Kim, M.G. Paper-based nucleic acid testing system for simple and early diagnosis of mosquito-borne RNA viruses from human serum. Biosens. Bioelectron. 2020, 151, 111998.
- Mitchell, S.L.; George, K.S. Evaluation of the COVID19 ID NOW EUA assay. J. Clin. Virol. 2020, 128, 104429.
- Zhuang, J.; Yin, J.; Lv, S.; Wang, B.; Mu, Y. Advanced “lab-on-a-chip” to detect viruses-Current challenges and future perspectives. Biosens. Bioelectron. 2020, 163, 112291.
- Van Tan, L.; Hong, N.T.T.; Ngoc, N.M.; Thanh, T.T.; Lam, V.T.; Nguyet, L.A.; Nhu, L.N.T.; Ny, N.T.H.; Minh, N.N.Q.; Man, D.N.H.; et al. SARS-CoV-2 and co-infections detection in nasopharyngeal throat swabs of COVID-19 patients by metagenomics. J. Infect. 2020, 81, e175–e177.
- Chandler, J.C.; Bevins, S.N.; Ellis, J.W.; Linder, T.J.; Tell, R.M.; Jenkins-Moore, M.; Root, J.J.; Lenoch, J.B.; Robbe-Austerman, S.; DeLiberto, T.J.; et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). bioRxiv 2021, 118, e2114828118.
- Yan, H.; Jiao, H.; Liu, Q.; Zhang, Z.; Xiong, Q.; Wang, B.-J.; Wang, X.; Guo, M.; Wang, L.-F.; Lan, K.; et al. ACE2 receptor usage reveals variation in susceptibility to SARS-CoV and SARS-CoV-2 infection among bat species. Nat. Ecol. Evol. 2021, 5, 600–608.
- Edwards, C.E.; Yount, B.L.; Graham, R.L.; Leist, S.R.; Hou, Y.J.; Dinnon, K.H.; Sims, A.C.; Swanstrom, J.; Gully, K.; Scobey, T.D.; et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. USA 2020, 117, 26915–26925.
- Pickering, B.; Smith, G.; Pinette, M.; Embury-Hyatt, C.; Moffat, E.; Marszal, P.; Lewis, C.E. Susceptibility of domestic swine to experimental infection with SARS-CoV-2. bioRxiv 2020, 27, 104–112.
- Santini, J.M.; Edwards, S.J. Host range of SARS-CoV-2 and implications for public health. Lancet Microbe 2020, 1, e141–e142.
- Wu, J.L.; Tseng, W.P.; Lin, C.H.; Lee, T.F.; Chung, M.Y.; Huang, C.H.; Chen, S.Y.; Hsueh, P.R.; Chen, S.C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020, 81, 435–442.
- Van Elslande, J.; Houben, E.; Depypere, M.; Brackenier, A.; Desmet, S.; André, E.; Van Ranst, M.; Lagrou, K.; Vermeersch, P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1082–1087.
- Roy, V.; Fischinger, S.; Atyeo, C.; Slein, M.; Loos, C.; Balazs, A.; Luedemann, C.; Astudillo, M.G.; Yang, D.; Wesemann, D.R.; et al. SARS-CoV-2-specific ELISA development. J. Immunol. Methods 2020, 484–485, 112832.
- Fang, W.H.; Yang, J.R.; Lin, C.Y.; Hsiao, P.J.; Tu, M.Y.; Chen, C.F.; Tsai, D.J.; Su, W.; Huang, G.S.; Chang, H.; et al. Accuracy augmentation of body composition measurement by bioelectrical impedance analyzer in elderly population. Medicine 2020, 99, e19103.
- Mak, G.C.; Cheng, P.K.; Lau, S.S.; Wong, K.K.; Lau, C.S.; Lam, E.T.; Chan, R.C.; Tsang, D.N.C. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 129, 104500.
- Goode, J.A.; Rushworth, J.V.; Millner, P.A. Biosensor Regeneration: A Review of Common Techniques and Outcomes. Langmuir 2015, 31, 6267–6276.
- Samson, R.; Navale, G.R.; Dharne, M.S. Biosensors: Frontiers in rapid detection of COVID-19. Biotech 2020, 10, 385.
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121.
- Xu, L.; Li, D.; Ramadan, S.; Li, Y.; Klein, N. Facile biosensors for rapid detection of COVID-19. Biosens. Bioelectron. 2020, 170, 112673.
This entry is offline, you can click here to edit this entry!
