Lentiviral-mediated integration of a CFTR transgene cassette into airway basal cells is a strategy being considered for cystic fibrosis (CF) cell-based therapies. However, CFTR expression is highly regulated in differentiated airway cell types and a subset of intermediate basal cells destined to differentiate. Since basal stem cells typically do not express CFTR, suppressing the CFTR expression from the lentiviral vector in airway basal cells may be beneficial for maintaining their proliferative capacity and multipotency. We identified miR-106b as highly expressed in proliferating airway basal cells and extinguished in differentiated columnar cells. Herein, we developed lentiviral vectors with the miR-106b-target sequence (miRT) to both study miR-106b regulation during basal cell differentiation and detarget CFTR expression in basal cells. Given that miR-106b is expressed in the 293T cells used for viral production, obstacles of viral genome integrity and titers were overcome by creating a 293T-B2 cell line that inducibly expresses the RNAi suppressor B2 protein from flock house virus. While miR-106b vectors effectively detargeted reporter gene expression in proliferating basal cells and following differentiation in the air–liquid interface and organoid cultures, the CFTR-miRT vector produced significantly less CFTR-mediated current than the non-miR-targeted CFTR vector following transduction and differentiation of CF basal cells. These findings suggest that miR-106b is expressed in certain airway cell types that contribute to the majority of CFTR anion transport in airway epithelium.
- miRNA
- airway basal cell
- CFTR
- gene therapy
- lentivirus
Note:All the information in this draft can be edited by authors. And the entry will be online only after authors edit and submit it.
1. Introduction
Cystic fibrosis (CF) is an inherited disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene [1]. CFTR is expressed primarily in epithelial cells of multiple organs. CFTR plays an important role in transepithelial anion transport important for regulating airway surface fluid volume, viscosity, and pH [2]. Lung disease with CF involves thick viscous mucus and chronic bacterial infections and is the primary cause of mortality. Gene and cell-based therapies for CF lung disease are gaining momentum, but knowledge gaps do remain regarding the target airway cell types that can prevent or reverse lung disease once a functional CFTR gene is expressed [3].
Both the proximal and distal airways express CFTR, but the landscape of cell types and CFTR expression patterns differ in these two levels of the airway. In the proximal airways, basal cells are considered the major stem cell precursor for ciliated cells, goblet cells, ionocytes, and other specialized cell types [3,4]. CFTR is expressed at widely divergent levels in a subset of proximal airway basal cells, secretory (goblet) cells, and ionocytes [5,6]. In the distal airway, basal and club cells are generally considered multipotent or bipotent stem cells, respectively, and can both give rise to ciliated cells. CFTR is most abundantly expressed in club secretory cells of bronchioles and alveolar type II cells [3,7,8].
Delivery of the CFTR gene to the CF airway basal cell is of particular interest in CF cell-based therapies, as this stem cell target has the ability to self-renew and differentiate into secretory cells (goblet or club), ciliated cells, and ionocytes. Lentiviral vectors have advantages over other widely used gene delivery vectors, such as adeno-associated vector (AAV), because lentiviruses integrate into the host genome and persist following cell division. However, CFTR is not typically expressed in multipotent airway basal cells but is rather expressed in transitional (intermediate) basal cells fated to become secretory cells [3,6,7]. Given that the functional role of CFTR expression in basal cell differentiation is unknown, methods to regulate transgene-derived CFTR expression in multipotent and transitional basal cell states and mimic endogenous patterns of expression could provide greater efficacy in CF cell therapy approaches.
We hypothesized that this pattern of expression could be achieved by suppressing CFTR expressionin multipotent basal cells via miRNA-mediated silencing. This approach of suppressing transgeneexpression in a specific cell type is most often referred to as “detargeting”. To this end, we sought to identify a miRNA that was selectively expressed in multipotent basal cells and identified miR-106b. Here, we describe the challenges and solutions for vector production using this approach, the analysis of dual reporter gene vectors that demonstrate the efficiency of basal cell detargeting of transgene expression, and the functional consequences of downregulating CFTR expression in CF human basal cells by assessing their capacities for generating CFTR currents following differentiation. We believe these vectors created will provide new opportunities for studying pathways that control lineage-commitment of airway basal cells, understanding cell type-specific functions of CFTR function, and ultimately aid in developing more effective gene therapy approaches for CF.
2. Basal Cells Stably Express miR-106b in Conditional Reprogramming Proliferative Cultures for Long-Term Culture
To select a miRNA for detargeting experiments, we accessed publicly available data through NCBI Gene Expression Omnibus (GEO) under serial number GSE22145 that compared basal cells vs. columnar cells in nasal airway [14] and found seven miRNAs that were consistently expressed in basal cells but not columnar cells from the nasal epithelia of three donors (Figure 1A). To evaluate the expression of miRNA expression in our cultured human tracheobronchial basal cells expanded in SAGM-EA [9], we used a TaqMan low-density array (TLDA; Applied Biosystems) to quantify relative expression of 377 miRNAs (Supplemental Table S1). Expression of 252 miRNAs was consistently detected in basal cells at passage 3 and at passage 18 (Figure 1B). Of these miRNAs, only nine changed significantly between passage 3 and 18 (FDR test with Q = 5%) and 171 miRNAs did not exceed a ± 2-fold change in expression in the passage (Figure 1C). Using a less stringent test, expression of 15 miRNAs changed significantly between passage 3 and 18 (unadjusted t-test p ≤ 0.05 and absolute fold change ≥ 2) (Figure 1D).
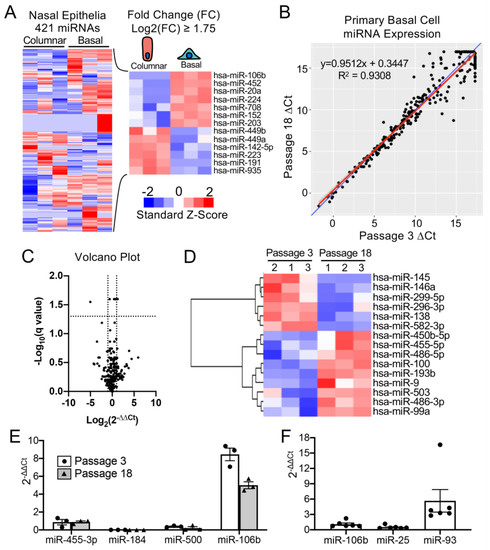
Figure 1. mMiR-106b is stably expressed at high levels in proliferating human basal cells. (A) Published data of miRNAs detected by high throughput sequence profiling of nasal basal cells and columnar cells (Accession: GSE22145) were used to generate a heatmap of 421 expressed miRNAs (left) and 13 miRNAs with a Log2 fold difference greater than 1.75 or less than −1.75 (right). (B) Correlation of miRNA expression in basal cells at Passage 3 and Passage 18 detected by qPCR array with the blue line represents a theoretical perfect correlation (x = y), and the red line represents linear regression model. (C) Volcano plot of miRNA array data indicating genes that were differentially expressed between passages 3 and 18. (D) Heatmap of miRNA array data with unsupervised hierarchical clustering of 15 miRNAs (of 252 detected) with an absolute fold change ≥ 2 and an unadjusted p value of ≤0.05. (E) Relative quantification of candidate basal cell-specific miRNAs, miR-184, miR-500, and miR-106b compared to a known basal cell-specific miR-455-3p. Freshly isolated primary human tracheobronchial cells were passaged 3 (P3) and 18 (P18) times in SAGM-EA media (N = 3). (F) Relative quantification of miRNAs belonging to miR-106b-25 cluster in passage 3 basal cells (N = 6). Each dot represents one donor.
Comparison of our array data with the nasal miRNA sequencing study demonstrated that miR-106b was one of the few miRNAs that was not expressed in columnar cells. Other miRNAs that were basal cell-specific in the nasal study included miR-184 and miR-500. miR-500 was detected at lower levels than miR-106b in our array study and miR-184 was undetectable. In this regard, miR-106b appeared to be the ideal miRNA to use in basal cell detargeting. We decided that our candidate miRNA should have a higher expression level than that of miR-455-3p, which has been reported to effectively inhibit MUC1 in human epithelial basal cells [15]. To more quantitively evaluate the expression of miR-106 in reference to low (miR-500) and very low (miR-184) basal cell expressing miRNAs, we performed single-plex qPCR for these miRNAs in comparison to that of miR-455-3p (Figure 1E). As expected, the expression levels of miR-184 expression was very low and miR-500a was absent, while miR-106b was more than 11-fold higher than the level of miR-455-3p. Moreover, miR-106b was stable on passage, decreasing by only 30% during the 15 passages. These findings confirmed the validity of the array data and suggested miR-106b was a top candidate for basal cell detargeting.
miR-106b, miR-25 and miR-93 belong to the miR-106b-25 cluster that is located in the 13th intron of mini-chromosome maintenance complex component 7 gene (MCM7) [16,17]. We prepared miRNA samples from six donors and analyzed the relative expression of these miRNAs (Figure 1F). The expression levels were fairly consistent between the six random donor samples. Notably, although these miRNAs are in the same cistron, their expression varied over a 10-fold range in airway basal cells (Figure 1F), and the pattern of expression of each of the three miRNAs was also different than that reported for the miR-106b-25 cluster in other tissues [18,19,20,21,22]. Although miR-93 was expressed at ~4.5-fold higher levels than miR-106b, we chose to move forward with miR-106b since miR-93 was observed to be expressed in differentiated human nasal columnar cells [14] (GEO dataset: GSE22145).
3. Increasing the Production Yield of a Lentiviral Vector Harboring Bidirectional Expression Cassettes
In order to evaluate detargeting using a basal cell-specific miR-target (miRT) site, we sought to have two reporter genes (one detargeted and one constitutively expressed) within the lentiviral vector. Since the miRT must reside in the 3′ UTR of the targeted gene cassette, creating this vector required two transgene cassettes (each with unique promoters and 3′ UTRs) oriented in the opposite direction (Figure 2A). We chose a nuclear-targeted EGFP (EGFP-nls) and Tomato as the two transgenes, with the miRT harbored in the 3′ UTR of the EGFP-nls cassette in the reverse orientation. The Tomato transgene in the direct orientation utilized the 3′-LTR polyA site and could not accommodate a miRT without compromising the viral packaging. This vector platform, we call LV-dt/EGFP, was constructed to allow the flexible insertion of any miRT sequence for specific cell type detargeting of transgene expression.
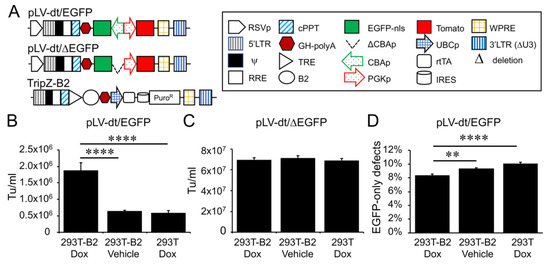
Figure 2. Suppressor of RNAi B2 protein increases titer and viral genome integrity of lentiviral vectors harboring bidirectional gene expression cassettes. (A) pLV-dt/ΔEGFP and pLV-dt/EGFP are the proviral vector plasmids used in production of these lentiviral vectors. TripZ-B2 is a lentiviral vector used to make the 293T-B2 cell line that expresses B2 following doxycycline treatment. The box legend to the right highlights the components of these proviral plasmids. Definitions are as follows: 5’LTR, 5’ long terminal repeat; ψ, psi, viral packaging signal sequence; RRE, rev response element, where Rev protein binds; cPPT, central polypurine tract, recognition site for proviral DNA synthesis; STOP, translation stop sequence; EGFP-nls, EGFP with nuclear localization signal; ΔCBAp, deletion of chicken beta-actin promoter (CBA); CBAp, CBA promoter in reverse orientation to the viral genomic transcript; PGKp, mouse phosphoglycerate kinase 1 promoter; TRE, tetracycline response element; UBCp, ubiquitin C promoter; rtTA, reverse tetracycline trans-activator; IRES, internal ribosomal entry site; WPRE, Woodchuck hepatitis virus post-transcriptional regulatory element for increasing nuclear export; 3’LTR (ΔU3), 3’ long terminal repeat with deletion in unique 3’ sequence that is necessary for activating viral genome transcription. (B) Comparison of LV-dt/EGFP titers produced by 293T-B2 with doxycycline treatment, 293T-B2 with vehicle treatment, and 293T with doxycycline treatment. Data show the mean+/-SEM for N = 6 viral preparations. (C) Comparison of lentiviral vector LV-dt/ΔEGFP transduction unit per milliliter (Tu/mL) produced by 293T-B2 with doxycycline treatment, 293T-B2 with vehicle (water) treatment, and 293T with doxycycline treatment. Data show the mean+/-SEM for N = 6 viral preparations. (D) Comparison of the percentage of cells positive for EGFP only among LV-dt/EGFP infected cells produced by 293T-B2 with doxycycline treatment, 293T-B2 with vehicle treatment, and 293T with doxycycline treatment. Data show the mean+/-SEM for N = 6 viral preparations. (B,C) Statistical comparisons were made by one-way ANOVA, Bonferroni’s multiple comparison test. ****, p < 0.0001. **, p < 0.01.
Initial attempts to generate the LV-dt/EGFP virus gave rise to low titers, despite lacking miRT sequences. We hypothesized complementarity of antisense EGFP mRNA, expressed from the proviral plasmid following transfection, with the full-length sense-strand viral RNA genome might activate RNAi and degrade viral genomes prior to packaging. To approach this problem, we sought to suppress RNAi during packaging with the flock house virus protein B2, which is a known RNAi suppressor [23,24,25]. When a B2-expression plasmid was co-transfected when making LV-dt/EGFP, the resulting virus titer was ~3 times higher than that without B2 (data not shown). We then used a lentivector to stably integrate a B2 gene expression cassette into 293T cells, however, persistent B2 expression in 293T cells was toxic. Thus, we generated a 293T cell line that expresses a doxycycline inducible (Tet-on) B2 protein using a TRIPZ vector (Figure 2A). We first tested different concentrations of doxycycline and two time points of doxycycline addition, at the time of proviral vector transfection or at the first media change after transfection. We observed that addition of 500 ng/mL doxycycline at the time of transfection produced highest virus titer while maintaining health of the producer cells. B2 mRNA induction by doxycycline was verified by qPCR (data not shown). Indeed, the lentiviral vector LV-dt/EGFP titer was significantly increased by doxycycline induced B2 expression during the virus production (Figure 2B). To confirm that the mechanism of reduced titers of LV-dt/EGFP was due to antisense EGFP transcripts, we created a second control vector (LV-dt/ΔEGFP) which lacked the CBA-promoter controlling EGFP expression. Titers of LV-dt/ΔEGFP, which lacked expression of EGFP transcripts with complementary to the viral genome, were not affected by the induction of B2 (Figure 2C).
To calculate viral titers in the above experiments, we used titration transduction assays on 293 cells followed by flow cytometry. We noticed that there were LV-dt/EGFP transduced cells that were positive for only EGFP or Tomato. This suggested that mutations or deletions within the proviral genomes likely occurred prior to packaging. During reverse transcription, a reverse transcriptase may change its templates 8 to 10 times [26] contributing to diversity of the lentivirus in the wild. This is a drawback to lentiviral vectors. We sought to evaluate whether inhibiting RNAi pathways with B2 would improve integrity of the packaged LV-dt/EGFP genomes. To this end, we compared the percentage of LV-dt/EGFP transduced 293T cells that only expressed EGFP (defective particles) from three types of viral preparation conditions: 1) 293T-B2 cells induced with doxycycline, 2) 293T-B2 cell not induced with doxycycline, and 3) 293T cells induced with doxycycline. Results from these flow cytometry comparisons demonstrated that group-1 and group-2 had ~20% and ~10% fewer defective particles than group-3, respectively (Figure 2D). We hypothesize that low level expression of B2 in the uninduced group-2 viral preparations improved integrity of the viral genomes when compared to 293T preparations lacking B2 (group-3).
4. Detargeting EGFP Expression in Proliferating Basal Cells.
To test whether the miR-106b target sequence (miRT) could be used to effectively detarget gene expression in basal cells, we generated a lentivirus vector that contained a nuclear targeted EGFP with the 3′-UTR miR-106b target sequence (pLV-dt/EGFP-miRT) and a control lentivirus vector with the miR-106bT sequence in reverse orientation (pLV-dt/EGFP-RmiRT; Figure 3A). We infected human epithelial basal cells grown in SAGM-EA with LV-dt/EGFP-miRT or LV-dt/EGFP-RmiRT and analyzed EGFP and Tomato expression by flow cytometry and fluorescent imaging. As hypothesized, the nuclei of the LV-dt/EGFP-RmiRT transduced basal cells were EGFP-positive, whereas basal cells transduced with the LV-dt/EGFP-miRT vector were EGFP-negative (Figure 3B). Thus, the miR-106b target sequence in the 3′-UTR appeared to successfully detarget EGFP expression in basal cells. To verify that miR-106b was indeed responsible for EGFP knock-down, we transfected FACS isolated Tomato-positive LV-dt/EGFP-miRT transduced cells with a miR-106b inhibitor. As expected, the LV-dt/EGFP-miRT transduced cells transfected with the miR-106b inhibitor recovered nuclear EGFP expression, while the mock transfected negative control cells did not (Figure 3C). The quantification of EGFP-positive only (Q4), Tomato-positive only (Q1), EGFP/Tomato-double-positive (Q2) and double-negative (Q3) cells are shown in the quadrants generated by flow cytometry (Figure 3D).
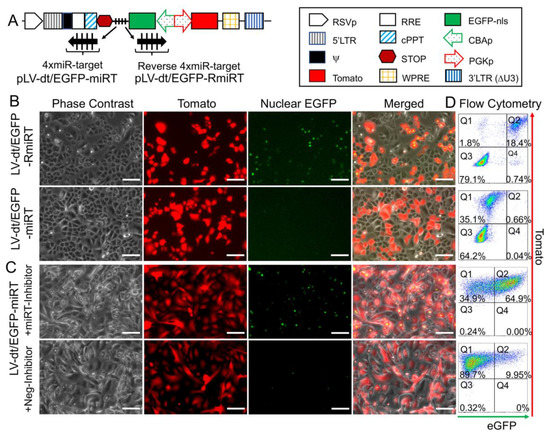
Figure 3. Incorporation of miR-106b target sequence (miRT) into the 3′-UTR of EGFP effectively detargets lentiviral-mediated expression in proliferating basal cells. (A) Diagram of the bidirectional promoter proviral lentiviral plasmids (pLV-dt/EGFP-miRT and pLV-dt/EGFP-RmiRT) used to generate lentivirus and test detargeting in basal cells. The box legend to the right highlights the components of these proviral plasmids as described in detail within the Figure 2A legend. LV-dt/EGFP-miRT is the experimental vector harboring a CBA promoter driven nuclear targeted EGFP (EGFP-nls) with miR-106b target sequence (4× miR target or mirT) in the reverse orientation. In the forward direction, the PGK promoter drives expression of the Tomato reporter, which is unaffected by miR-106b. LV-dt/EGFP-RmiRT is a control vector with the miRNA target sequence in the reverse orientation. (B) LV-dt/EGFP-miRT and LV-dt/EGFP-RmiRT viruses were used to transduce primary human airway basal cell in SAGM-EA cultures. The Tomato-positive (red) cells indicate the virally transduced cells. EGFP expression is seen in dt/EGFP-RmiRT control transduced cells but not in cells transduced with the detargeted LV-dt/EGFP-miRT vector. Scale bar, 100 µm. (C) Basal cells transduced with LV-dt/EGFP-miRT vector, and FACS isolated for Tomato-positive cells, were transfected with miRT-106b inhibitor sequences to block detargeting or mock transfected. Scale bar, 100 µm. (D) The cells in (B and C) were analyzed by flow cytometer and are shown in dot plots to the right of the corresponding images for each condition. The percentage of cells are indicated in each quadrant: Q1 (Tomato-positive only cells), Q2 (Tomato and EGFP double-positive cells), Q3 (EGFP-positive only cells), and Q4 (non-fluorescent cells).
5. Basal Cell miRT-106b Detargeting is Partially Maintained in Differentiated ALI Cultures and Organoids
miR-106 is highly expressed in proliferating basal cells grown in SAGM-EA media and basal cell detargeting with miRT-106b is highly effective (Figure 3). To determine if miR-106 expression in basal cells of differentiated cultures was sufficient for detargeting, we studied the EGFP expression profiles of ALI and organoid cultures generated from LV-dt/EGFP-miRT and LV-dt/EGFP-RmiRT transduced basal cells. Approximately 40% of the basal cell population was transduced and the cells were not subjected to FACS prior to making ALI cultures or airway organoids. ALI cultures generated from these two groups were sectioned and evaluated for EGFP, Tomato, and KRT5 (basal cell marker) expression. Both LV-dt/EGFP-miRT and LV-dt/EGFP-RmiRT transduced basal cells formed a pseudostratified epithelium with Tomato expression marking transduced cells and KRT5 marking the basal cell layer (Figure 4). As expected, LV-dt/EGFP-miRT transduced cultures lacked nuclear EGFP expression in the majority of KRT5-positive basal cells, confirming detargeting, but contained EGFP-positive nuclei in differentiated columnar cells (Figure 4A). Conversely, LV-dt/EGFP-RmiRT transduced control cultures contained nuclear EGFP-positive cells throughout the basal layer as well as the columnar cell differentiated layer (Figure 4B). Notably, LV-dt/EGFP-miRT transduced ALI culture lacked EGFP in ~50% of columnar cells, whereas the vast majority of columnar cells expressed EGFP in LV-dt/EGFP-RmiRT cultures. These findings suggest that miR106b may also be expressed or retained in a subset of columnar cells in ALI cultures.
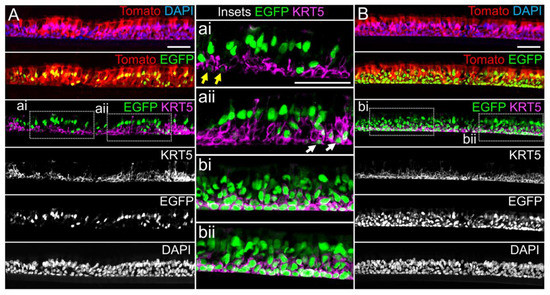
Figure 4. Lentiviral-mediated basal cell detargeting of EGFP following differentiation at an air–liquid interface. Primary human basal cells were transduced with LV-dt/EGFP-miRT or LV-dt/EGFP-RmiRT vectors and expanded for 1–2 days before seeding for differentiation in air–liquid interface (ALI) cultures. (A,B) Confocal microscopic images of (A) LV-dt/EGFP-miRT and (B) LV-dt/EGFP-RmiRT transduced ALI culture using sections immunostained for KRT5 (keratin-5) and imaged for KRT5, EGFP, and Tomato expression with DAPI to mark nuclei. Dual and single channel images are shown. Enlarged boxed regions in (A,B) are shown in the middle column. Scale bar, 50 μm.
We next performed similar studies in airway organoid cultures generated from LV-dt/EGFP-miRT and LV-dt/EGFP-RmiRT transduced basal cells (Figure 5A). These airway organoids mature with time to form an external basal cell layer and internal luminal cell layer composed of differentiated columnar cells. Similar to ALI cultures, confocal imaging of intact organoids demonstrated that LV-dt/EGFP-RmiRT transduced organoids contained EGFP expressing cells that spanned the outer basal cell layer as well as the differentiated luminal cell layer. By contrast, Tomato-positive LV-dt/EGFP-miRT transduced cells of the organoid lacked EGFP expression in the outer basal cell layer, but EGFP-positive luminal cells were observed. Tomato expression was dimmer than in the outer layer of basal cells in both groups, similar to ALI cultures, suggesting that the PGK promoter may be less active in the basal cells.
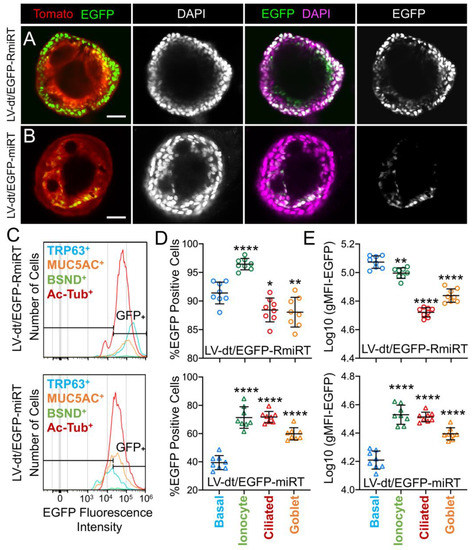
Figure 5. Basal cell detargeting of a reporter transgene in differentiated cell types of ALI cultures. (A,B) Human basal cells were transduced with (A) LV-dt/EGFP-RmiRT or (B) LV-dt/EGFP-miRT and expanded for 1–2 days prior to seeding in organoid culture. Confocal microscopic images of live organoids stained with the Hoescht 33342 nuclei marker. Single and dual channel images are pseudocolored to better project nuclear EGFP expression. Scale bar, 50μm. (C–E) Primary basal cells were transduced with LV-dt/EGFP-RmiRT or LV-dt/EGFP-miRT viruses and then FACS was used to isolated pure Tomato-positive basal cells. These cells were expanded in culture and then seeded into ALI cultures for differentiation and then detached and immunostained for quantification of EGFP expression in various cells types by flow cytometer. (C) Epithelial lineages were stained for TRP63/p63 (basal cells; blue), BSND (ionocytes; green), alpha-tubulin (ciliated), and MUC5AC (goblet cells; orange). Representative histogram distributions of lineage-labeled cell populations treated transduced with LV-dt/EGFP-RmiRT (top) or LV-dt/EGFP-miRT (bottom). (D) Percentage of EGFP-positive cells for each lineage using the gate shown in (C) which captures 90% of EGFP-positive basal cells in the control LV-dt/EGFP-RmiRT vector group. (E) Mean fluorescent intensity (MFI) of lineage-labeled populations. Statistics represent a one-way ANOVA with Dunnett’s multiple comparison test against the basal cell population: * p < 0.01, ** p < 0.05, **** p < 0.0001. Data show the mean +/-SD for N = 8 transwells for each condition.
To quantify the extent of detargeting in basal cells, we transduced primary human basal cells with LV-dt/EGFP-miRT or LV-dt/EGFP-RmiRT vector systems and FACS isolated Tomato-positive cells for expansion in SAGM-EA prior to seeding into ALI cultures. Well-differentiated ALI cultures were then dissociated, and the single cell suspension of epithelial cells was fixed and stained for markers of basal cells (TRP63), ciliated cells (acetylated tubulin), goblet cells (MUC5AC), and ionocytes (BSND). These populations were then subjected to flow cytometer and the percentages of EGFP-positive cells for each cell phenotype quantified (Figure 5C–E). As expected from confocal imaging of ALI cultures (Figure 4), the fluorescent intensity of all cell types in the LV-dt/EGFP-miRT group was lower than that of LV-dt/EGFP-RmiRT, suggesting that inclusion of the miRT-106b target sequences generally reduces expression of the EGFP transgene. However, quantification of the percentage of EGFP-positive cells demonstrated the largest drop for miRT vs. RmiRT expression in TRP63-positive basal cells (2.3-fold) (Figure 5D). Furthermore, in LV-dt/EGFP-RmiRT transduced cells, the percentage of EGFP-positive basal cells was significantly lower than ciliated and goblet cells, whereas the opposite was observed in LV-dt/EGFP-miRT transduced cells (Figure 5D). Additionally, the mean fluorescent intensity (MFI, calculated as the geometric mean) of EGFP was the highest in basal cells of the LV-dt/EGFP-RmiRT control group, supporting confocal imaging of ALI demonstrating the strongest EGFP expression in KRT5-positive basal cells with this vector (Figure 4B, bi, bii). By contrast, the MFI was the lowest in the LV-dt/EGFP-miRT transduced basal cells as compared to ionocytes, ciliated cells and goblet cells (Figure 5E), similar to those observed histologic studies (Figure 4A, ai, aii). Overall, these findings suggest that the miRT-106b sequences effectively reduce expression of EGFP in basal cells.
An unexpected finding from these cellular phenotyping studies of LV-dt/EGFP-miRT- and LV-dt/EGFP-RmiRT-transduced epithelia was a significant shift in the number of goblet cells and ionocytes (Table 1). The largest shift occurred in the percentage of MUC5AC-positive goblet cells, rising 2-fold (p < 0.0001) in LV-dt/EGFP-miRT transduced epithelia as compared to the RmiRT control vector. By contrast, the percentage of ionocytes marginally declined in the LV-dt/EGFP-miRT group (p < 0.0388), while the percentage of basal cells and ciliated cells was not significantly different between the two groups. These findings raise the interesting possibility that high-level expression of mRNA containing the miRT sequence could potentially sequester miR-106b and impact processes involved in goblet cell and ionocyte specification.
Table 1. Distribution of cell types in differentiated ALI cultures.
| Vector | % Basal Cells (TRP63+) | % Ionocytes (BSND+) | % Ciliated Cells (Ac-Tubulin+) | % Goblet Cells (MUC5AC+) |
|---|---|---|---|---|
| LV-dt/EGFP-RmiRT | 21.8+/−3.8 * | 0.82+/−0.02 | 44.3+/−1.6 | 11.2+/−0.8 |
| LV-dt/EGFP-miRT | 22.4+/−1.5 | 0.72+/−0.04 | 41.5+/−0.9 | 21.9+/−1.2 |
| p-value ** | 0.7768 | 0.0388 | 0.1567 | <0.0001 |
6. Basal Cell-Detargeting of CFTR Expression Alters Functional Complementation in CF Airway Epithelia
To test our primary hypothesis that detargeting of CFTR in basal cells would improve complementation in CF airway epithelia, we replaced EGFP in LV-dt/EGFP-miRT with CFTR to generate the pLV-dt/CFTR-miRT lentiviral vector. Our control vector (pLV-dt/CFTR-Ø) was identical to pLV-dt/CFTR-miRT but lacked the miR-106b target sequences (Figure 6A). Freshly isolated CF human tracheobronchial basal cells were transduced with each vector and expanded 4 days before seeding into transwells for ALI culture. Contrary to our initial hypothesis, ALI cultures transduced with LV-dt/CFTR-Ø gave rise to ~3.5-fold greater CFTR-mediated CI– currents than that of LV-dt/CFTR-miRT transduced ALI cultures (Figure 6B), even though both cultures expressed similar levels of CFTR mRNA, which were 3.2-fold (LV-dt/CFTR-Ø) and 2.7-fold (LV-dt/CFTR-miRT) higher levels than the mock-infected group. Characteristic of CFTR, these currents were induced by cAMP agonists (IBMX/Forskolin) and inhibited by the CFTR channel blocker GlyH101. The slightly lower expression of CFTR mRNA in the LV-dt/CFTR-miRT transduced cultures was expected, consistent with detargeted expression in basal cells.
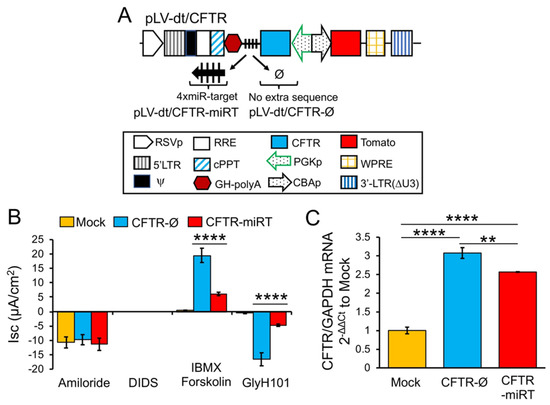
Figure 6. Detargeting CFTR expression in basal cells impacts the level of complementation in CF airway epithelia. (A) Diagram of lenti-vector containing CFTR expression cassette in reverse orientation. The PGK promoter (PGKp) drives expression of CFTR with the miR-106b target sequence (4× miR target or mirT) in the 3’UTR. CBA promoter drives expression of Tomato as a reporter gene for viral transduction. pLV-dt/CFTR-Ø is a control vector with no miRT sequence. Box (below) is a legend for each shape in the diagram that highlights the components of these proviral plasmids as described in detail within the Figure 2A legend. (B) Short-circuit current (Isc) measurements of differentiated air-liquid interface cultures seeded with transduced at basal cells. Mock, mock-infected cells. PGK-CFTR-Ø, cells transduced by LV-dt/CFTR-Ø; PGK-CFTR-miRT, cells transduced by LV-dt/CFTR-miRT. Amiloride was used to block ENaC-mediated Na+ currents. 4,4′-Diisothiocyanatostilbene-2,2′-disulfonic acid (DIDS) was use to inhibit most non-CFTR chloride channel. 3-isobutyl-2-methylxanthine (IBMX) and Forskolin was used for activate CFTR channels. N-(2-naphtalenyl)-(3.5-dibromo-2.4-dihydroxyphenyl)methylene glycine hydrazide (GlyH101) was used to block CFTR. Data show the mean +/-SEM for N = 6 transwells for each condition. (C) Relative quantification of CFTR mRNA normalized to GAPDH mRNA from each sample used in B. For B and C, the statistics used is one-way ANOVA, Tukey’s multiple comparisons test. ****, p < 0.0001. **, p = 0.0025. Data show the mean +/-SEM for N = 3 independent samples for each condition.
References
- Riordan, J.R.; Rommens, J.M.; Kerem, B.; Alon, N.; Rozmahel, R.; Grzelczak, Z.; Zielenski, J.; Lok, S.; Plavsic, N.; Chou, J.L.; et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 1989, 245, 1066–1073.
- Xie, Y.; Ostedgaard, L.; Abou Alaiwa, M.H.; Lu, L.; Fischer, A.J.; Stoltz, D.A. Mucociliary Transport in Healthy and Cystic Fibrosis Pig Airways. Ann. Am. Thorac. Soc. 2018, 15, S171–S176.
- Tang, Y.; Yan, Z.; Engelhardt, J.F. Viral Vectors, Animal Models, and Cellular Targets for Gene Therapy of Cystic Fibrosis Lung Disease. Hum. Gene Ther. 2020, 31, 524–537.
- Rock, J.R.; Randell, S.H.; Hogan, B.L. Airway basal stem cells: A perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 2010, 3, 545–556.
- Montoro, D.T.; Haber, A.L.; Biton, M.; Vinarsky, V.; Lin, B.; Birket, S.E.; Yuan, F.; Chen, S.; Leung, H.M.; Villoria, J.; et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018, 560, 319–324.
- Plasschaert, L.W.; Zilionis, R.; Choo-Wing, R.; Savova, V.; Knehr, J.; Roma, G.; Klein, A.M.; Jaffe, A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018, 560, 377–381.
- Carraro, G.; Mulay, A.; Yao, C.; Mizuno, T.; Konda, B.; Petrov, M.; Lafkas, D.; Arron, J.R.; Hogaboam, C.M.; Chen, P.; et al. Single Cell Reconstruction of Human Basal Cell Diversity in Normal and IPF Lung. Am. J. Respir. Crit. Care Med. 2020.
- Xu, Y.; Mizuno, T.; Sridharan, A.; Du, Y.; Guo, M.; Tang, J.; Wikenheiser-Brokamp, K.A.; Perl, A.T.; Funari, V.A.; Gokey, J.J.; et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 2016, 1, e90558.
- Mou, H.; Vinarsky, V.; Tata, P.R.; Brazauskas, K.; Choi, S.H.; Crooke, A.K.; Zhang, B.; Solomon, G.M.; Turner, B.; Bihler, H.; et al. Dual SMAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell Stem Cell 2016.
- Barde, I.; Salmon, P.; Trono, D. Production and titration of lentiviral vectors. Curr. Protoc. Neurosci. 2010, 53, 4–21.
- Denning, W.; Das, S.; Guo, S.; Xu, J.; Kappes, J.C.; Hel, Z. Optimization of the transductional efficiency of lentiviral vectors: Effect of sera and polycations. Mol. Biotechnol. 2013, 53, 308–314.
- Sun, X.; Olivier, A.K.; Liang, B.; Yi, Y.; Sui, H.; Evans, T.I.; Zhang, Y.; Zhou, W.; Tyler, S.R.; Fisher, J.T.; et al. Lung phenotype of juvenile and adult cystic fibrosis transmembrane conductance regulator-knockout ferrets. Am. J. Respir. Cell Mol. Biol. 2014, 50, 502–512.
- Yan, Z.; Sun, X.; Feng, Z.; Li, G.; Fisher, J.T.; Stewart, Z.A.; Engelhardt, J.F. Optimization of Recombinant Adeno-Associated Virus-Mediated Expression for Large Transgenes, Using a Synthetic Promoter and Tandem Array Enhancers. Hum. Gene Ther. 2015, 26, 334–346.
- Marcet, B.; Chevalier, B.; Luxardi, G.; Coraux, C.; Zaragosi, L.E.; Cibois, M.; Robbe-Sermesant, K.; Jolly, T.; Cardinaud, B.; Moreilhon, C.; et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 2011, 13, 693–699.
- Martinez-Anton, A.; Sokolowska, M.; Kern, S.; Davis, A.S.; Alsaaty, S.; Taubenberger, J.K.; Sun, J.; Cai, R.; Danner, R.L.; Eberlein, M.; et al. Changes in microRNA and mRNA expression with differentiation of human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 2013, 49, 384–395.
- Mehlich, D.; Garbicz, F.; Wlodarski, P.K. The emerging roles of the polycistronic miR-106b approximately 25 cluster in cancer-A comprehensive review. Biomed Pharmacother. 2018, 107, 1183–1195.
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783.
- Zhou, Y.; Hu, Y.; Yang, M.; Jat, P.; Li, K.; Lombardo, Y.; Xiong, D.; Coombes, R.C.; Raguz, S.; Yague, E. The miR-106b~25 cluster promotes bypass of doxorubicin-induced senescence and increase in motility and invasion by targeting the E-cadherin transcriptional activator EP300. Cell Death Differ. 2014, 21, 462–474.
- Chuang, T.D.; Luo, X.; Panda, H.; Chegini, N. miR-93/106b and their host gene, MCM7, are differentially expressed in leiomyomas and functionally target F3 and IL-8. Mol. Endocrinol. 2012, 26, 1028–1042.
- Haldar, S.; Roy, A.; Banerjee, S. Differential regulation of MCM7 and its intronic miRNA cluster miR-106b-25 during megakaryopoiesis induced polyploidy. RNA Biol. 2014, 11, 1137–1147.
- Kan, T.; Sato, F.; Ito, T.; Matsumura, N.; David, S.; Cheng, Y.; Agarwal, R.; Paun, B.C.; Jin, Z.; Olaru, A.V.; et al. The miR-106b-25 Polycistron, Activated by Genomic Amplification, Functions as an Oncogene by Suppressing p21 and Bim. Gastroenterology 2009, 136, 1689–1700.
- Smith, A.L.; Iwanaga, R.; Drasin, D.J.; Micalizzi, D.S.; Vartuli, R.L.; Tan, A.C.; Ford, H.L. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene 2012, 31, 5162–5171.
- Chao, J.A.; Lee, J.H.; Chapados, B.R.; Debler, E.W.; Schneemann, A.; Williamson, J.R. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 2005, 12, 952–957.
- Li, H.; Li, W.X.; Ding, S.W. Induction and suppression of RNA silencing by an animal virus. Science 2002, 296, 1319–1321.
- Lingel, A.; Simon, B.; Izaurralde, E.; Sattler, M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005, 6, 1149–1155.
- Rawson, J.M.O.; Nikolaitchik, O.A.; Keele, B.F.; Pathak, V.K.; Hu, W.S. Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity. Nucleic. Acids Res. 2018, 46, 10535–10545.
- Rock, J.R.; Onaitis, M.W.; Rawlins, E.L.; Lu, Y.; Clark, C.P.; Xue, Y.; Randell, S.H.; Hogan, B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 12771–12775.
This entry is adapted from the peer-reviewed paper 10.3390/genes11101169
