In post-mortem studies, a significant dysregulation of electron transport chain (ETC) complexes was observed in patients with neurodegenerative diseases (NDs). These findings strongly implicate that mitochondrial dysfunction-linked alterations in oxidative phosphorylation (OXPHOS) can be considered a highly relevant molecular mechanism in different NDs. Histopathological examinations revealed decreased complex I level, preferentially in the substantia nigra (SN), in patients suffering from Parkinson’s disease (PD). These findings are consistent with the fact that inhibitors of complex I (such as the environmental toxins MPTP or rotenone) can cause parkinsonism in animal models and humans. Huntington’s disease (HD) has been associated with defects of complex II and, to a lesser extent, complex IV. The chronic administration of the complex II inhibitor 3-nitropropionic acid causes an HD-like phenotype in rodent and non-human primate models. In Alzheimer’s disease (AD), widespread cortical complex IV defects were identified in post-mortem brain tissue. The in vivo neuroimaging-based assessment of electron transport chain (ETC)-related metabolite levels could thus help elucidate the complex role of OXPHOS disturbances in NDs.
- OXPHOS-Related Complexes and Metabolites
- Neuroimaging
1. 31Phosphorus-Magnetic Resonance Spectroscopy Imaging to Quantify Oxidative Phosphorylation (OXPHOS)-Related Metabolite Levels In Vivo
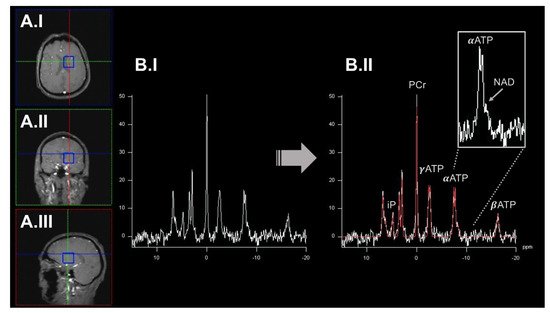
2. Dynamic Measurements of OXPHOS Reaction Kinetics by 31phosphorus Magnetization Transfer Magnetic Resonance Spectroscopy Imaging
3. Quantitative Assessment of Mitochondrial Complex I by Positron Emission Tomography Imaging Radiotracer
4. Broadband Near-Infrared Spectroscopy to Dynamically Map Cytochrome c Oxidase Activity
Abbreviations
(b)NIRS: (broadband) near-infrared spectroscopy imaging. (ox)CCO: oxidized cytochrome c oxidase. 18F-BCPP-EF: 2-tert-butyl-4-chloro-5-(6-(2-(2(18F)fluoroethoxy) -ethoxy]-pyridin-3-ylmethoxy)-2H-pyridazin-3-one. AD: Alzheimer’s disease. ADP: adenosine diphosphate. AMP: adenosine monophosphate. APD: atypical parkinsonism. ASL: arterial spin labeling. ATP: adenosine triphosphate. DNA: deoxyribonucleic acid. ETC: electron transport chain. HD: Huntington’s disease. HEP: high-energy phosphorus-containing metabolites. iP: inorganic phosphate. MND: motor neuron disease. MPTP: 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. MRI: magnetic resonance imaging. MRSI: magnetic resonance spectroscopy imaging. MT-MRSI: magnetization transfer magnetic resonance spectroscopy imaging. mtDNA: mitochondrial DNA. NAD/NAD+/NADH: nicotinamide adenine dinucleotide. ND: neurodegenerative diseases. NMR: nuclear magnetic resonance. OEF: oxygen extraction fraction. OXPHOS: oxidative phosphorylation. PD: Parkinson’s disease. PET: positron emission tomography. QUEST-MRI: QUEnch-assiSTed-MRI. ROS: reactive oxygen species. SN: substantia nigra. SNR: signal-to-noise ratio. TCA: tricarboxylic acid.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23137263
References
- Zhu, X.H.; Lu, M.; Chen, W. Quantitative imaging of brain energy metabolisms and neuroenergetics using in vivo X-nuclear 2H, 17O and 31P MRS at ultra-high field. J. Magn. Reson. 2018, 292, 155–170.
- Buonocore, M.H.; Maddock, R.J. Magnetic resonance spectroscopy of the brain: A review of physical principles and technical methods. Rev. Neurosci. 2015, 26, 609–632.
- Liu, Y.; Gu, Y.; Yu, X. Assessing tissue metabolism by phosphorous-31 magnetic resonance spectroscopy and imaging: A methodology review. Quant. Imaging Med. Surg. 2017, 7, 707–726.
- Das, N.; Ren, J.; Spence, J.; Chapman, S.B. Phosphate Brain Energy Metabolism and Cognition in Alzheimer’s Disease: A Spectroscopy Study Using Whole-Brain Volume-Coil 31Phosphorus Magnetic Resonance Spectroscopy at 7Tesla. Front. Neurosci. 2021, 15, 641739.
- Rango, M.; Dossi, G.; Squarcina, L.; Bonifati, C. Brain mitochondrial impairment in early-onset Parkinson’s disease with or without PINK1 mutation. Mov. Disord. 2020, 35, 504–507.
- Stamelou, M.; Pilatus, U.; Reuss, A.; Magerkurth, J.; Eggert, K.M.; Knake, S.; Ruberg, M.; Schade-Brittinger, C.; Oertel, W.H.; Hoglinger, G.U. In vivo evidence for cerebral depletion in high-energy phosphates in progressive supranuclear palsy. J. Cereb. Blood Flow Metab. 2009, 29, 861–870.
- Mochel, F.; N’Guyen, T.M.; Deelchand, D.; Rinaldi, D.; Valabregue, R.; Wary, C.; Carlier, P.G.; Durr, A.; Henry, P.G. Abnormal response to cortical activation in early stages of Huntington disease. Mov. Disord. 2012, 27, 907–910.
- Lu, M.; Zhu, X.H.; Zhang, Y.; Chen, W. Intracellular redox state revealed by in vivo (31) P MRS measurement of NAD(+) and NADH contents in brains. Magn. Reson. Med. 2014, 71, 1959–1972.
- Lu, M.; Zhu, X.H.; Chen, W. In vivo (31) P MRS assessment of intracellular NAD metabolites and NAD(+) /NADH redox state in human brain at 4 T. NMR Biomed. 2016, 29, 1010–1017.
- Reiten, O.K.; Wilvang, M.A.; Mitchell, S.J.; Hu, Z.; Fang, E.F. Preclinical and clinical evidence of NAD(+) precursors in health, disease, and ageing. Mech. Ageing Dev. 2021, 199, 111567.
- Chong, R.; Wakade, C.; Seamon, M.; Giri, B.; Morgan, J.; Purohit, S. Niacin Enhancement for Parkinson’s Disease: An Effectiveness Trial. Front. Aging Neurosci. 2021, 13, 667032.
- Du, F.; Zhu, X.H.; Qiao, H.; Zhang, X.; Chen, W. Efficient in vivo 31P magnetization transfer approach for noninvasively determining multiple kinetic parameters and metabolic fluxes of ATP metabolism in the human brain. Magn. Reson. Med. 2007, 57, 103–114.
- Adanyeguh, I.M.; Rinaldi, D.; Henry, P.G.; Caillet, S.; Valabregue, R.; Durr, A.; Mochel, F. Triheptanoin improves brain energy metabolism in patients with Huntington disease. Neurology 2015, 84, 490–495.
- van den Bogaard, S.J.; Dumas, E.M.; Teeuwisse, W.M.; Kan, H.E.; Webb, A.; van Buchem, M.A.; Roos, R.A.; van der Grond, J. Longitudinal metabolite changes in Huntington’s disease during disease onset. J. Huntingtons Dis. 2014, 3, 377–386.
- Harada, N.; Nishiyama, S.; Kanazawa, M.; Tsukada, H. Development of novel PET probes, BCPP-EF, BCPP-BF, and BCPP-EM for mitochondrial complex 1 imaging in the living brain. J. Labelled Comp. Radiopharm. 2013, 56, 553–561.
- Tsukada, H.; Nishiyama, S.; Fukumoto, D.; Kanazawa, M.; Harada, N. Novel PET probes 18F-BCPP-EF and 18F-BCPP-BF for mitochondrial complex I: A PET study in comparison with 18F-BMS-747158-02 in rat brain. J. Nucl. Med. 2014, 55, 473–480.
- Tsukada, H.; Ohba, H.; Kanazawa, M.; Kakiuchi, T.; Harada, N. Evaluation of 18F-BCPP-EF for mitochondrial complex 1 imaging in the brain of conscious monkeys using PET. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 755–763.
- Tsukada, H.; Kanazawa, M.; Ohba, H.; Nishiyama, S.; Harada, N.; Kakiuchi, T. PET Imaging of Mitochondrial Complex I with 18F-BCPP-EF in the Brains of MPTP-Treated Monkeys. J. Nucl. Med. 2016, 57, 950–953.
- Wilson, H.; Pagano, G.; de Natale, E.R.; Mansur, A.; Caminiti, S.P.; Polychronis, S.; Middleton, L.T.; Price, G.; Schmidt, K.F.; Gunn, R.N.; et al. Mitochondrial Complex 1, Sigma 1, and Synaptic Vesicle 2A in Early Drug-Naive Parkinson’s Disease. Mov. Disord. 2020, 35, 1416–1427.
- Terada, T.; Therriault, J.; Kang, M.S.P.; Savard, M.; Pascoal, T.A.; Lussier, F.; Tissot, C.; Wang, Y.T.; Benedet, A.; Matsudaira, T.; et al. Mitochondrial complex I abnormalities is associated with tau and clinical symptoms in mild Alzheimer’s disease. Mol. Neurodegener. 2021, 16, 28.
- Mansur, A.; Rabiner, E.A.; Tsukada, H.; Comley, R.A.; Lewis, Y.; Huiban, M.; Passchier, J.; Gunn, R.N. Test-retest variability and reference region-based quantification of (18)F-BCPP-EF for imaging mitochondrial complex I in the human brain. J. Cereb. Blood Flow Metab. 2021, 41, 771–779.
- Bale, G.; Elwell, C.E.; Tachtsidis, I. From Jobsis to the present day: A review of clinical near-infrared spectroscopy measurements of cerebral cytochrome-c-oxidase. J. Biomed. Opt. 2016, 21, 091307.
- Lange, F.; Dunne, L.; Hale, L.; Tachtsidis, I. MAESTROS: A Multiwavelength Time-Domain NIRS System to Monitor Changes in Oxygenation and Oxidation State of Cytochrome-C-Oxidase. IEEE J. Sel. Top. Quantum Electron. 2019, 25, 7100312.
- Kovacsova, Z.; Bale, G.; Mitra, S.; de Roever, I.; Meek, J.; Robertson, N.; Tachtsidis, I. Investigation of Confounding Factors in Measuring Tissue Saturation with NIRS Spatially Resolved Spectroscopy. Adv. Exp. Med. Biol. 2018, 1072, 307–312.
- Caldwell, M.; Scholkmann, F.; Wolf, U.; Wolf, M.; Elwell, C.; Tachtsidis, I. Modelling confounding effects from extracerebral contamination and systemic factors on functional near-infrared spectroscopy. Neuroimage 2016, 143, 91–105.
- Russell-Buckland, J.; Kaynezhad, P.; Mitra, S.; Bale, G.; Bauer, C.; Lingam, I.; Meehan, C.; Avdic-Belltheus, A.; Martinello, K.; Bainbridge, A.; et al. Systems Biology Model of Cerebral Oxygen Delivery and Metabolism During Therapeutic Hypothermia: Application to the Piglet Model. Adv. Exp. Med. Biol. 2021, 1269, 31–38.
