Amaryllidaceae alkaloids (AAs) are plant specialized metabolites with therapeutic properties exclusively produced by the Amaryllidaceae plant family. Bioengineered microbial hosts that grow rapidly can produce plant target specialized metabolites faster as compared to whole plant systems. In addition, the production of plant metabolites in heterologous hosts can reduce downstream extraction process, which eventually becomes more economically sustainable. For the successful synthesis of plant metabolites such as AAs, heterologous hosts require the introduction of reconstructed biosynthetic pathway, requiring key enzymes. This requires comprehensive knowledge of the enzymatic reactions involved in the biosynthesis of the compound of interest in the native host organisms (i.e., plants).
- amaryllidaceae alkaloids
- bioactive molecules
- biotechnological approach
- biosynthesis
- in vitro cultures
- synthetic biology
1. Molecular Understanding of Amaryllidaceae Alkaloids Biosynthesis
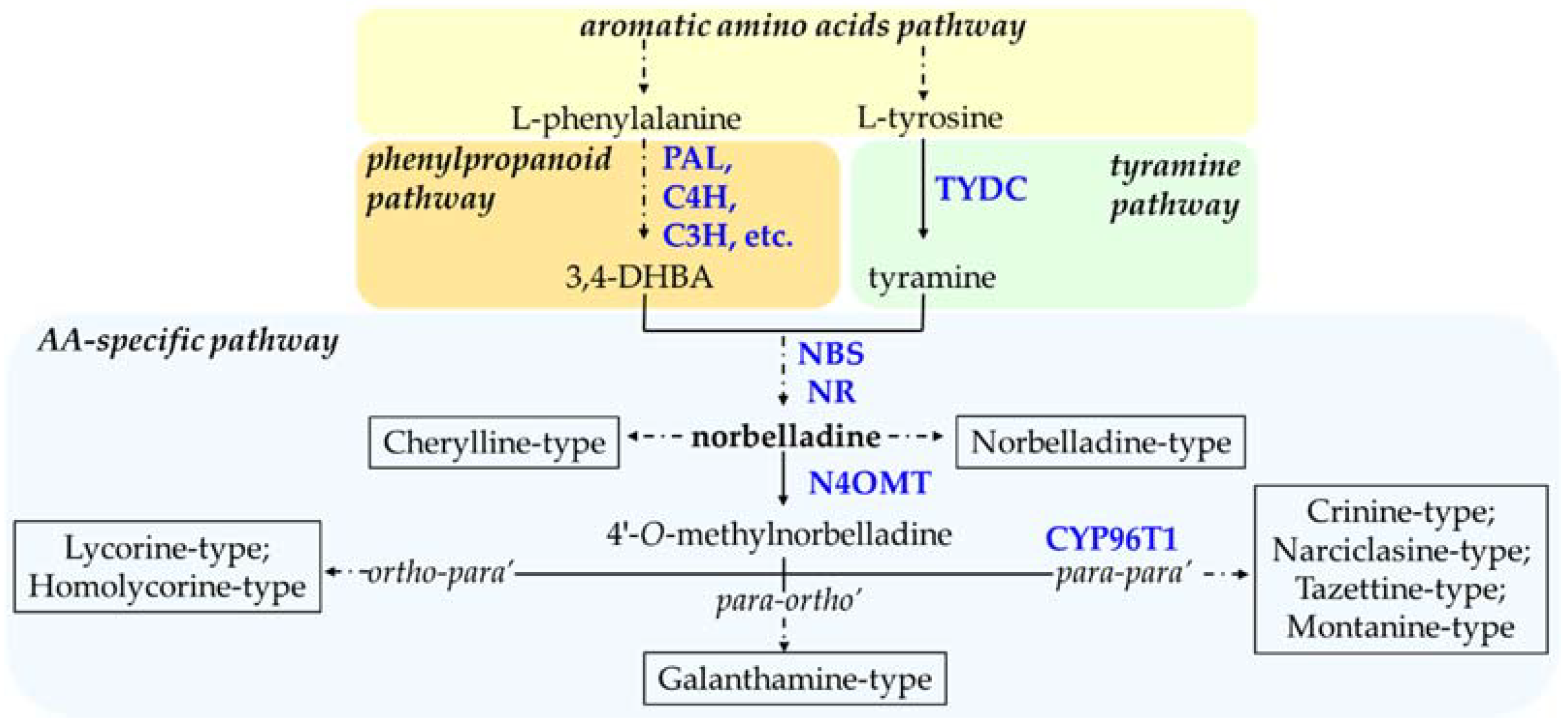
2. Synthetic Biology for AA Biosynthesis
This entry is adapted from the peer-reviewed paper 10.3390/biom12070893
References
- Desgagné-Penix, I. Biosynthesis of alkaloids in Amaryllidaceae plants: A review. Phytochem. Rev. 2020, 20, 409–431.
- Kilgore, M.B.; Kutchan, T.M. The Amaryllidaceae alkaloids: Biosynthesis and methods for enzyme discovery. Phytochem. Rev. Proc. Phytochem. Soc. Eur. 2016, 15, 317–337.
- Hotchandani, T.; de Villers, J.; Desgagne-Penix, I. Developmental Regulation of the Expression of Amaryllidaceae Alkaloid Biosynthetic Genes in Narcissus papyraceus. Genes 2019, 10, 594.
- Li, Q.; Xu, J.; Zheng, Y.; Zhang, Y.; Cai, Y. Transcriptomic and Metabolomic Analyses Reveals That Exogenous Methyl Jasmonate Regulates Galanthamine Biosynthesis in Lycoris longituba Seedlings. Front. Plant Sci. 2021, 12, 713795.
- Singh, A.; Desgagne-Penix, I. Transcriptome and metabolome profiling of Narcissus pseudonarcissus ‘King Alfred’ reveal components of Amaryllidaceae alkaloid metabolism. Sci. Rep. 2017, 7, 17356.
- Tousignant, L.; Diaz-Garza, A.M.; Majhi, B.B.; Gelinas, S.E.; Singh, A.; Desgagne-Penix, I. Transcriptome analysis of Leucojum aestivum and identification of genes involved in norbelladine biosynthesis. Planta 2022, 255, 30.
- Kilgore, M.B.; Augustin, M.M.; Starks, C.M.; O’Neil-Johnson, M.; May, G.D.; Crow, J.A.; Kutchan, T.M. Cloning and Characterization of a Norbelladine 4′-O-Methyltransferase Involved in the Biosynthesis of the Alzheimer’s Drug Galanthamine in Narcissus sp. aff. pseudonarcissus. PLoS ONE 2014, 9, e103223.
- Park, C.H.; Yeo, H.J.; Park, Y.E.; Baek, S.A.; Kim, J.K.; Park, S.U. Transcriptome Analysis and Metabolic Profiling of Lycoris Radiata. Biology 2019, 8, 63.
- Hu, J.; Li, W.; Liu, Z.; Zhang, G.; Luo, Y. Molecular cloning and functional characterization of tyrosine decarboxylases from galanthamine-producing Lycoris radiata. Acta Physiol. Plantarum 2021, 43, 84.
- Li, Q.; Xu, J.; Yang, L.; Zhou, X.; Cai, Y.; Zhang, Y. Transcriptome Analysis of Different Tissues Reveals Key Genes Associated With Galanthamine Biosynthesis in Lycoris longituba. Front. Plant Sci. 2020, 11, 519752.
- Singh, A.; Massicotte, M.A.; Garand, A.; Tousignant, L.; Ouellette, V.; Berube, G.; Desgagne-Penix, I. Cloning and characterization of norbelladine synthase catalyzing the first committed reaction in Amaryllidaceae alkaloid biosynthesis. BMC Plant Biol. 2018, 18, 338.
- Kilgore, M.B.; Holland, C.K.; Jez, J.M.; Kutchan, T.M. Identification of a Noroxomaritidine Reductase with Amaryllidaceae Alkaloid Biosynthesis Related Activities. J. Biol. Chem. 2016, 291, 16740–16752.
- Majhi, B.B.; Gélinas, S.-E.; Mérindol, N.; Desgagne-Penix, I. Characterization of norbelladine synthase and noroxomaritidine/norcraugsodine reductase reveals a novel catalytic route for the biosynthesis of Amaryllidaceae alkaloids including the Alzheimer’s drug galanthamine. Plant J. 2022; submitted.
- Kilgore, M.B.; Augustin, M.M.; May, G.D.; Crow, J.A.; Kutchan, T.M. CYP96T1 of Narcissus sp. aff. pseudonarcissus Catalyzes Formation of the Para-Para’ C-C Phenol Couple in the Amaryllidaceae Alkaloids. Front. Plant Sci. 2016, 7, 225.
- Ncube, B.; Nair, J.J.; Rárová, L.; Strnad, M.; Finnie, J.F.; Van Staden, J. Seasonal pharmacological properties and alkaloid content in Cyrtanthus contractus N.E. Br. S. Afr. J. Botany 2015, 97, 69–76.
- Katoh, A.; Ohki, H.; Inai, K.; Hashimoto, T. Molecular regulation of nicotine biosynthesis. Plant Biotechnol. 2005, 22, 389–392.
- Onoyovwe, A.; Hagel, J.M.; Chen, X.; Khan, M.F.; Schriemer, D.C.; Facchini, P.J. Morphine biosynthesis in opium poppy involves two cell types: Sieve elements and laticifers. Plant Cell 2013, 25, 4110–4122.
- El Tahchy, A.; Ptak, A.; Boisbrun, M.; Barre, E.; Guillou, C.; Dupire, F.; Chretien, F.; Henry, M.; Chapleur, Y.; Laurain-Mattar, D. Kinetic study of the rearrangement of deuterium-labeled 4′-O-methylnorbelladine in Leucojum aestivum shoot cultures by mass spectrometry. Influence of precursor feeding on amaryllidaceae alkaloid accumulation. J. Nat. Prod. 2011, 74, 2356–2361.
- Ren, Z.; Lin, Y.; Lv, X.; Zhang, J.; Zhang, D.; Gao, C.; Wu, Y.; Xia, Y. Clonal bulblet regeneration and endophytic communities profiling of Lycoris sprengeri, an economically valuable bulbous plant of pharmaceutical and ornamental value. Sci. Hortic. 2021, 279, 109856.
- Ka, S.; Masi, M.; Merindol, N.; Di Lecce, R.; Plourde, M.B.; Seck, M.; Gorecki, M.; Pescitelli, G.; Desgagne-Penix, I.; Evidente, A. Gigantelline, gigantellinine and gigancrinine, cherylline- and crinine-type alkaloids isolated from Crinum jagus with anti-acetylcholinesterase activity. Phytochemistry 2020, 175, 112390.
- Fennell, C.W.; Elgorashi, E.E.; van Staden, J. Alkaloid production in Crinum moorei cultures. J. Nat. Prod. 2003, 66, 1524–1526.
- Aleya, F.; Xianmin, C.; Anthony, H.; Meriel, J. Relative expression of putative genes involved in galanthamine and other Amaryllidaceae alkaloids biosynthesis in Narcissus field and in vitro tissues. Gene 2021, 774, 145424.
- Sun, B.; Wang, P.; Wang, R.; Li, Y.; Xu, S. Molecular Cloning and Characterization of a meta/para-O-Methyltransferase from Lycoris aurea. Int. J. Mol. Sci. 2018, 19, 1911.
- Li, Y.; Li, S.; Thodey, K.; Trenchard, I.; Cravens, A.; Smolke, C.D. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. USA 2018, 115, E3922–E3931.
- Diamond, A.; Desgagne-Penix, I. Metabolic engineering for the production of plant isoquinoline alkaloids. Plant Biotechnol. J. 2016, 14, 1319–1328.
- Facchini, P.J.; Bohlmann, J.; Covello, P.S.; De Luca, V.; Mahadevan, R.; Page, J.E.; Ro, D.K.; Sensen, C.W.; Storms, R.; Martin, V.J. Synthetic biosystems for the production of high-value plant metabolites. Trends Biotechnol. 2012, 30, 127–131.
- Fossati, E.; Narcross, L.; Ekins, A.; Falgueyret, J.P.; Martin, V.J. Synthesis of Morphinan Alkaloids in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0124459.
- Cao, M.; Gao, M.; Suástegui, M.; Mei, Y.; Shao, Z. Building microbial factories for the production of aromatic amino acid pathway derivatives: From commodity chemicals to plant-sourced natural products. Metab. Eng. 2020, 58, 94–132.
- Li, Y.; Li, J.; Qian, B.; Cheng, L.; Xu, S.; Wang, R. De Novo Biosynthesis of p-Coumaric Acid in E. coli with a trans-Cinnamic Acid 4-Hydroxylase from the Amaryllidaceae Plant Lycoris aurea. Molecules 2018, 23, 3185.
- Patnaik, R.; Zolandz, R.R.; Green, D.A.; Kraynie, D.F. L-tyrosine production by recombinant Escherichia coli: Fermentation optimization and recovery. Biotechnol. Bioeng. 2008, 99, 741–752.
- Slattery, S.S.; Diamond, A.; Wang, H.; Therrien, J.A.; Lant, J.T.; Jazey, T.; Lee, K.; Klassen, Z.; Desgagne-Penix, I.; Karas, B.J.; et al. An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 2018, 7, 328–338.
- Farhi, M.; Marhevka, E.; Ben-Ari, J.; Algamas-Dimantov, A.; Liang, Z.; Zeevi, V.; Edelbaum, O.; Spitzer-Rimon, B.; Abeliovich, H.; Schwartz, B.; et al. Generation of the potent anti-malarial drug artemisinin in tobacco. Nat. Biotechnol. 2011, 29, 1072–1074.
- Kitney, R.; Freemont, P. Synthetic biology—The state of play. FEBS Lett. 2012, 586, 2029–2036.
- Auslander, S.; Auslander, D.; Fussenegger, M. Synthetic Biology-The Synthesis of Biology. Angew. Chem. Int. Ed. Engl. 2017, 56, 6396–6419.
- Matasci, N.; Hung, L.H.; Yan, Z.; Carpenter, E.J.; Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Ayyampalayam, S.; Barker, M.; et al. Data access for the 1000 Plants (1KP) project. Gigascience 2014, 3, 17.
- Xiao, M.; Zhang, Y.; Chen, X.; Lee, E.J.; Barber, C.J.; Chakrabarty, R.; Desgagne-Penix, I.; Haslam, T.M.; Kim, Y.B.; Liu, E.; et al. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 2013, 166, 122–134.
- Pyne, M.E.; Narcross, L.; Martin, V.J.J. Engineering Plant Secondary Metabolism in Microbial Systems. Plant Physiol. 2019, 179, 844–861.
