Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Nucleosome-Omics is one of the subdisciplines of Omics, which studies nucleosome-level phenomenon on chromatin and genome 3D landscape, including the interaction and combination among histones, trancriptional factors and DNA, by combining nucleosome resolution omics technologies with high-throughput sequencing techniques.
- Nucleosome-Omics
- epigenetics
- nucleosomes
1. Introduction of Nucleosome-Omics
The nuclear genetic code of eukaryotes (such as animals and plants) depends on chromatin, which is a highly organized complex of DNA and proteins and is a principal component of the cell nucleus [1]. What is amazing is that chromatin almost carries most of the genetic information about organic development, growth, and aging. With conventional electron microscopy measurements, it was found that chromatin containing roughly 2 m DNA is condensed and folded in the nucleus, which is only about 10 μm in diameter [2]. The diameter of a classical chromatin fiber is just only ~30 nm [3]. Scientists have reached a consensus that nucleosomes could be thought of as the basic building blocks of chromatin [4]. In addition, the “beads on a string” conformation vividly depicts the state of nucleosomes in chromatin [5]. During the different biological processes, such as cell division, cell proliferation, and differentiation, the “beads on a string” nucleosomes can flexibly compress and loosen with DNA duplication and transcriptional regulation. Nucleosomes are basal structural units, which consist of histone proteins and organizers. They are assembled into compact structures called chromatin in interphase, or into enormous, high-order structures called chromosomes in mitosis [1]. Scientists have used classical physics tools and methods, electron microscopy, X-ray scattering and crystallography, etc., and found or proposed some models [3]. For example, from the 1970s–2010s, there are the one-start solenoid model [6], a two-start helical ribbon model [7], a two-start crossed-linker model [8], a polymer melt model [9], and a two-start zig-zag model with tetranucleosomal unit [10]. This makes the nucleosome folding structure and pattern more and more clear [3]. Therefore, in the early days of nucleosome research, especially twenty years ago, these studies mainly focused on ultrastructural observation and structural model analysis [3]. However, the effects of chromatin conformation and nucleosome dynamics on transcriptional regulation have been poorly studied and unclear. Recently, with the development of next-generation sequencing (NGS) technology, various nucleosome-resolution or even base-pair resolution-level genomics sequencing technologies have been developed in succession. For instance, based on nucleosome-level genome fragment digestion by micrococcal nuclease (MNase), MNase-seq and derivative technology [11], MNase-Exo-seq [12], MNase-ChIP-seq [13], MACC-seq [14], etc. was developed. With these technologies or integrating with other omics technologies (ChIP-seq, RNA-seq, or others), accurate nucleosome position, the distribution of specific genomic regions, and their impacts on transcriptional regulation and gene expression were revealed [15]. Fortunately, it is worth pointing out that the development of three-dimensional (3D) genomics technologies makes it possible to analyze the three-dimensional conformations of chromatin. Hi-C and ChIA-PET are the representative technologies that burst the field of epigenetic research [16]. By combining Hi-C with MNase genome fragmentation, Micro-C and its derivative Micro-C XL can be improved [17]. These technologies help unmask ultra-fine-scale chromatin ultrastructure at the 100–1000 base-pair resolution [18]. Some special structures, dots, and stripes, which are not easily found in the Hi-C, are well represented and reconstructed [19].
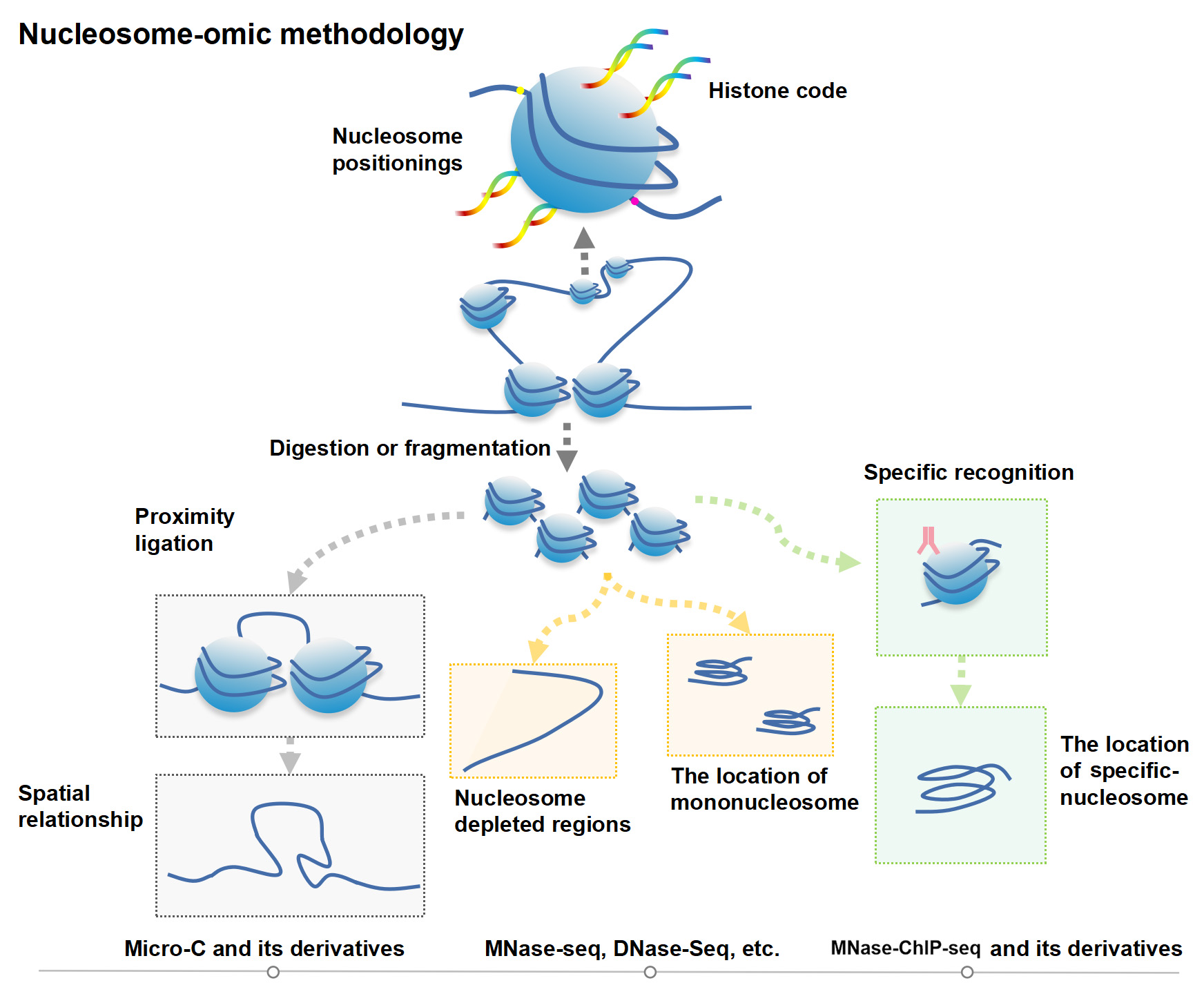 Figure 1. Nucleosome-moic methodology.
Figure 1. Nucleosome-moic methodology. 2. Extended information
2.1. Scientific Discovery, Development and Related Research of Nucleosomes
Chromatin is the carrier of genetic and epigenetic information about organisms. Under an optical microscope, at the 1400 nM length interval, researchers could observe some regular-shaped and compact chromosome packages called metaphase chromosomes, and sometimes, loose and seemingly irregular chromatin in other interphase cells [20]. The chromosome packages the genome via a hierarchical series of folding steps ranging from 700 nM, 300 nM, to 30 nM (Figure 2a) [6]. In addition, the chromatin can directly extend 30 nM chromatin fibrils (Figure 2a) [20]. Thirty nM chromatin fiber’s substructure and its higher-order structure are hot spots in the current spatial chromatin conformation epigenetics research [10]. Physical and biochemical approaches complement each other to study the chromatin spatial conformation [3]. By using microscopic imaging techniques, ChromEMT, the nucleus, chromatins, nucleosomes, and chromatin loops are visualized [20].
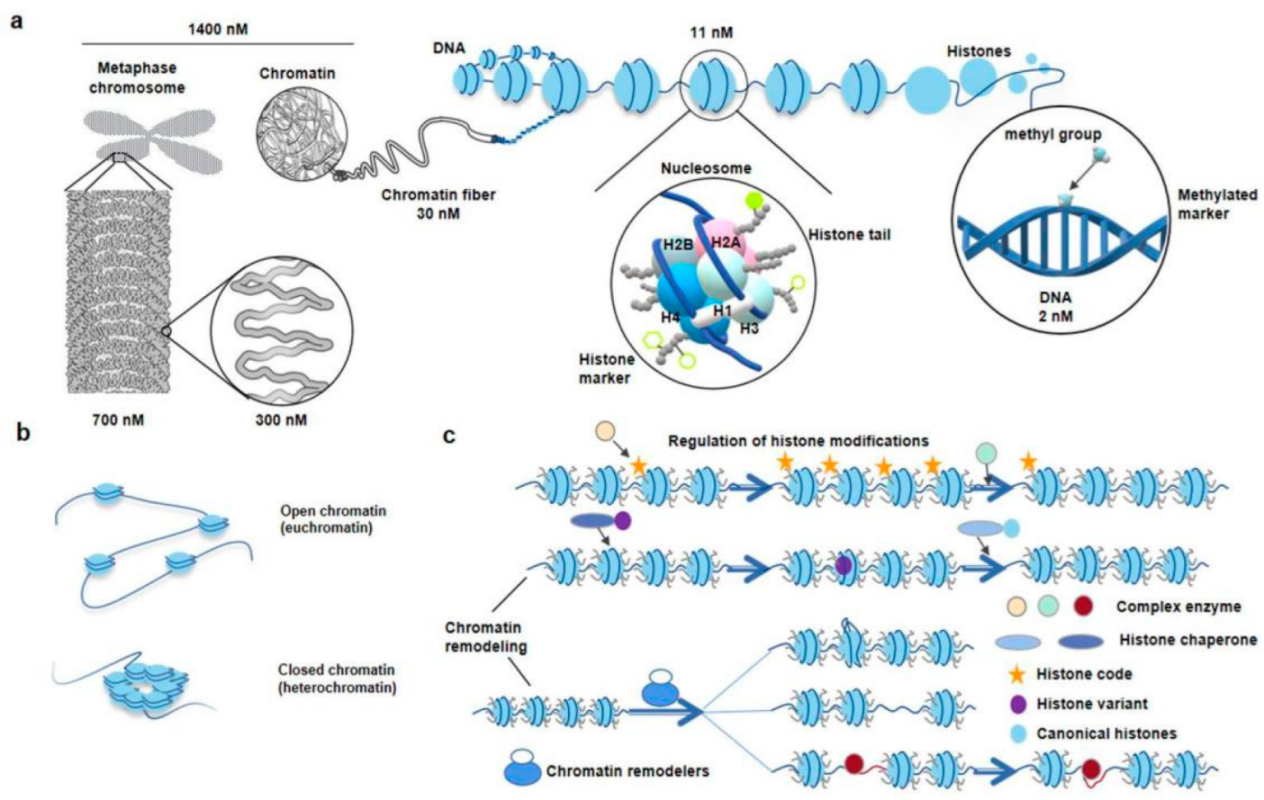
Figure 2. The diagram of hierarchical structures from chromatin to nucleosome in eukaryotes. (a), Zoom in chromosome or chromatin to DNA. The DNA double helix wraps around histone octamers to form nucleosomes, the basic structural units of chromatin. The classic histone octamer is made up of four types of histone (H2A/H2B/H3/H4) and has eight histone tails. Covalent modification markers on histone tails play an important role in regulating the chromatin structure and function. DNA methylation markers are also epigenetic codes. (b), Different states of chromatin. The regions of chromatin, called euchromatin, exist in an extended and loosely packed state, which is conducive to transcription. Conversely, densely packed heterochromatin is silent. (c), Regulation of histone modifications and chromatin remodeling. Recognition, reading, and removal of histone modifications depend on the Writer, Reader, and Eraser. The complex enzymes responsible for the alteration of histone modifications often have more than one ability. When the reading modules of these complexes bind to the corresponding sites, their writing or erasure modules are activated and work at nearby sites. ATP-driven chromatin remodelers, which are complex of multiple subunits, catalyze chromatin remodeling. Meanwhile, the nucleosome structure usually changes in four ways: replacement, dissociation, removing, and slide. Replacement indicates chromatin remodelers, which catalyze replacement between canonical histone and histone variants in nucleosomes. Dissociation indicates that the double helix DNA wrapped around the nucleosome loosens. Removing indicates the disintegration of nucleosomes. Slide indicates chromatin remodelers can allow nucleosomes to slide along DNA without unwinding the DNA double strand [21]. Courtesy: National Human Genome Research Institute (https://www.genome.gov/genetics-glossary/Chromatin, accessed on 19 May 2022; https://www.genome.gov/genetics-glossary/Nucleosome, accessed on 19 May 2022).
During the 1970s, the nucleosome and nucleosome “beads on a string” model were first discovered and proposed (Figure 2a) [22]. Nucleosomes are the basic structural units of eukaryotic chromatin, which is composed of DNA and histones. Under the 11 nM diameter sub-chromatin fiber, each nucleosome consists of 145–147 bp of DNA wrapped around 1.75 histone octamer loops, where the histone octamer consists of two copies of the four core histones H2A, H2B, H3, and H4. Histone isoforms H1 are a linker histone [23]. Nucleosomes control cellular processes by affecting transcription factor accessibility to DNA [24]. Aggregation and deaggregation of nucleosomes are consistent with closed chromatin (heterochromatin) and open chromatin (euchromatin) (Figure 2b), which are related to transcriptional repression and activation. When the genomic regions have little nucleosome occupancy and become nucleosome-depleted regions (NDRs), the protein factors (pre-transcriptional initiation complex, chromatin remodelers, complex enzymes, histone chaperone, histone variants, transcription factor, or RNA polymerase II will access the region and affect the transcriptional expression of nearby genes (Figure 2c) [25].
As is known, DNA methylation and histone modification are two important directions of classical epigenetic research (Figure 2a) [26][27]. They have important associations with gene function and expression patterns. The nucleosome position and histone components change are important parts of chromatin remodeling (Figure 2c) [28][29]. They are critical for gene expression and most DNA-level processes. Besides, nuclear phosphatidylinositol phosphates (PIPs) are also critical interactors and regulators of chromatin remodeling and transcription and thus are crucial for establishing a transcriptionally competent landscape of the cell nucleus [30][31]. The nucleosome position is affected by many factors, such as DNA sequence, transcription factor, RNA polymerase II, preinitiation complex, and ATP-dependent nucleosome remodeling enzymes [28]. Scientists cannot find any one factor alone that determines the nucleosome position. Sometimes, the histone components are not classical octameric units; the canonical histone is replaced with a histone variant during special biological processes and altered gene expression patterns [32]. Thus, the mobilization, disassembly, and destabilization of nucleosomes are new insights that can be a breakthrough for exploring the mechanism that influences gene expression regulation and other eukaryotic DNA processes [25].
2.2. Significance of Studying Chromatin Nucleosome and Nucleosome-Level DNA
The packaging of eukaryotic genome into chromatin affects every process that occurs on DNA. Different cells have different nucleosome organization characteristics, and nucleosomes can occupy specific locations in some cells but not in others [33]. The positioning of nucleosomes on the underlying DNA plays a key role in the regulation of these processes [28]. The presence or absence of nucleosomes in a gene expression regulatory region determines whether transcription factors can bind to that particular region [34]. Moreover, the arrangement of nucleosomes along the DNA sequence is also a regulatory mechanism that affects gene expression and other DNA-dependent processes [35]. Therefore, to understand gene regulation quantitatively, it is necessary to know not only the precise location of nucleosomes (i.e., nucleosome positioning), but also the percentage of cells that contain nucleosomes at a given location (i.e., nucleosome occupancy) [28].
With the development of next-generation sequencing technology, it is possible to accurately locate nucleosomes at the genome-wide level and calculate the nucleosome occupancy rate of the whole genome [36].
This entry is adapted from the peer-reviewed paper 10.3390/genes13071114
References
- Maloy, S.R.; Hughes, K.T. Brenner’s Encyclopedia of Genetics, 2nd ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2013.
- Goel, V.Y.; Hansen, A.S. The Macro and Micro of Chromosome Conformation Capture. Wiley Interdiscip. Rev. Dev. Biol. 2020, 10, e395.
- Ohno, M.; Priest, D.; Taniguchi, Y. Nucleosome-Level 3D Organization of the Genome. Biochem. Soc. Trans. 2018, 46, 491–501.
- Oudet, P.; Gross-Bellard, M.; Chambon, P. Electron Microscopic and Biochemical Evidence That Chromatin Structure Is A Repeating Unit. Cell 1975, 4, 281–300.
- Olins, A.L.; Olins, D.E. Spheroid Chromatin Units (V Bodies). Science 1974, 183, 330–332.
- Finch, J.T.; Klug, A. Solenoidal Model for Superstructure in Chromatin. Proc. Natl. Acad. Sci. USA 1976, 73, 1897–1901.
- Woodcock, C.L.; Frado, L.L.; Rattner, J.B. The Higher-Order Structure of Chromatin: Evidence for a Helical Ribbon Arrangement. J. Cell Biol. 1984, 99, 42–52.
- Williams, S.; Athey, B.; Muglia, L.; Schappe, R.; Gough, A.; Langmore, J. Chromatin Fibers are Left-Handed Double Helices with Diameter and Mass Per Unit Length That Depend on Linker Length. Biophys. J. 1986, 49, 233–248.
- Eltsov, M.; MacLellan, K.M.; Maeshima, K.; Frangakis, A.S.; Dubochet, J. Analysis of Cryo-Electron Microscopy Images Does Not Support the Existence of 30-Nm Chromatin Fibers In Mitotic Chromosomes In Situ. Proc. Natl. Acad. Sci. USA 2008, 105, 19732–19737.
- Song, F.; Chen, P.; Sun, D.; Wang, M.; Dong, L.; Liang, D.; Xu, R.-M.; Zhu, P.; Li, G. Cryo-EM Study of the Chromatin Fiber Reveals a Double Helix Twisted by Tetranucleosomal Units. Science 2014, 344, 376–380.
- Schwartz, U.; Németh, A.; Diermeier, S.; Exler, J.H.; Hansch, S.; Maldonado, R.; Heizinger, L.; Merkl, R.; Längst, G. Characterizing the Nuclease Accessibility of DNA in Human Cells to Map Higher Order Structures of Chromatin. Nucleic Acids Res. 2019, 47, 1239–1254.
- Ocampo, J.; Cui, F.; Zhurkin, V.B.; Clark, D.J. The Proto-Chromatosome: A Fundamental Subunit of Chromatin? Nucleus 2016, 7, 382–387.
- Zhang, Z.; Pugh, B.F. High-Resolution Genome-wide Mapping of the Primary Structure of Chromatin. Cell 2011, 144, 175–186.
- Mieczkowski, J.; Cook, A.; Bowman, S.K.; Mueller, B.; Alver, B.H.; Kundu, S.; Deaton, A.M.; Urban, J.A.; Larschan, E.; Park, P.J.; et al. MNase Titration Reveals Differences Between Nucleosome Occupancy And Chromatin Accessibility. Nat. Commun. 2016, 7, 11485.
- Yu, J.; Xiong, C.; Zhuo, B.; Wen, Z.; Shen, J.; Liu, C.; Chang, L.; Wang, K.; Wang, M.; Wu, C.; et al. Analysis of Local Chromatin States Reveals Gene Transcription Potential During Mouse Neural Progenitor Cell Differentiation. Cell Rep. 2020, 32, 108072.
- Kong, S.; Zhang, Y. Deciphering Hi-C: From 3D genome to function. Cell Biol. Toxicol. 2019, 35, 15–32.
- Hsieh, T.-H.S.; Weiner, A.; Lajoie, B.; Dekker, J.; Friedman, N.; Rando, O.J. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell 2015, 162, 108–119.
- Mozziconacci, J.; Koszul, R. Filling the Gap: Micro-C Accesses the Nucleosomal Fiber at 100-1000 Bp Resolution. Genome Biol. 2015, 16, 169.
- Chang, L.-H.; Noordermeer, D. Of Dots and Stripes: The Morse Code of Micro-C Reveals the Ultrastructure of Transcriptional and Architectural Mammalian 3D Genome Organization. Mol. Cell 2020, 78, 376–378.
- Ou, H.D.; Phan, S.; Deerinck, T.J.; Thor, A.; Ellisman, M.H.; O’Shea, C.C. ChromEMT: Visualizing 3D Chromatin Structure and Compaction in Interphase and Mitotic Cells. Science 2017, 357, eaag0025.
- Zhou, C.Y.; Johnson, S.L.; Gamarra, N.I.; Narlikar, G.J. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu. Rev. Biophys. 2016, 45, 153–181.
- Kornberg, R.D.; Thomas, J.O. Chromatin Structure: Oligomers of the Histones. Science 1974, 184, 865–868.
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal Structure of the Nucleosome Core Particle at 2.8 a Resolution. Nature 1997, 389, 251–260.
- Barbier, J.; Vaillant, C.; Volff, J.N.; Brunet, F.G.; Audit, B. Coupling between Sequence-Mediated Nucleosome Organization and Genome Evolution. Genes 2021, 12, 851.
- Henikoff, S. Nucleosome Destabilization in the Epigenetic Regulation of Gene Expression. Nat. Rev. Genet. 2008, 9, 15–26.
- Yang, Y.; Fan, X.; Yan, J.; Chen, M.; Zhu, M.; Tang, Y.; Liu, S.; Tang, Z. A Comprehensive Epigenome Atlas Reveals DNA Methylation Regulating Skeletal Muscle Development. Nucleic Acids Res. 2021, 49, 1313–1329.
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct Features of H3K4me3 and H3K27me3 Chromatin Domains in Pre-Implantation Embryos. Nature 2016, 537, 558–562.
- Struhl, K.; Segal, E. Determinants of Nucleosome Positioning. Nat. Struct. Mol. Biol. 2013, 20, 267–273.
- Tan, S.; Davey, C.A. Nucleosome Structural Studies. Curr. Opin. Struct. Biol. 2011, 21, 128–136.
- Hoboth, P.; Sztacho, M.; Šebesta, O.; Schätz, M.; Castano, E.; Hozák, P. Nanoscale Mapping of Nuclear Phosphatidylinositol Phosphate Landscape by Dual-Color dSTORM. Biochim. et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2021, 1866, 158890.
- Sztacho, M.; Salovska, B.; Cervenka, J.; Balaban, C.; Hoboth, P.; Hozak, P. Limited Proteolysis-Coupled Mass Spectrometry Identifies Phosphatidylinositol 4,5-Bisphosphate Effectors in Human Nuclear Proteome. Cells 2021, 10, 68.
- Sharma, A.B.; Dimitrov, S.; Hamiche, A.; Van Dyck, E. Centromeric and Ectopic Assembly of CENP-A Chromatin in Health and Cancer: Old Marks and New Tracks. Nucleic Acids Res. 2019, 47, 1051–1069.
- Eitoku, M.; Sato, L.; Senda, T.; Horikoshi, M. Histone Chaperones: 30 years from Isolation to Elucidation of the Mechanisms of Nucleosome Assembly and Disassembly. Cell. Mol. Life Sci. 2007, 65, 414–444.
- Kornberg, R.D. The Molecular Basis of Eucaryotic Transcription. Cell Death Differ. 2008, 14, 1989–1997.
- Tao, Y.; Zheng, W.; Jiang, Y.; Ding, G.; Hou, X.; Tang, Y.; Li, Y.; Gao, S.; Chang, G.; Zhang, X.; et al. Nucleosome Organizations in Induced Pluripotent Stem Cells Reprogrammed from Somatic Cells Belonging to Three Different Germ Layers. BMC Biol. 2014, 12, 109.
- Schones, D.E.; Cui, K.; Cuddapah, S.; Roh, T.-Y.; Barski, A.; Wang, Z.; Wei, G.; Zhao, K. Dynamic Regulation of Nucleosome Positioning in the Human Genome. Cell 2008, 132, 887–898.
This entry is offline, you can click here to edit this entry!
