Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Insulin-like growth factor two (IGF-II) is a key protein regulating growth, particularly during normal fetal development, but it is also often dysregulated during tumorigenesis. IGF-II belongs to a larger system including several different regulatory factors and generally referred to as the “IGF system”. IGF-I and IGF-II are the main ligands of the type 1 IGF receptor (IGF-1R).
- IGF2
- growth
- Parental imprinting
1. Main Characteristics, Linear Organization
In 2019, Baral et al. unraveled the complexity of the genomic and transcriptional organization of the IGF2/Igf2 locus, by using human or mouse DNA segments as queries in genome analyses, and RNA sequencing libraries (complete review in [1]). IGF2 (ENSG00000167244) is composed of 10 exons and five promoters, whereas its mouse counterpart is located on chromosome 7, and is composed of eight exons and four promoters.
The five human IGF2 promoters control the expression of different non-coding exons, but all transcripts include exons 8–10, which encode the IGF-II protein precursor and the 3′ untranslated RNA. Human promoters 1 and 2 (P1 and P2) are species specific, and P2 regulates two classes of IGF2 transcripts differing due to alternative splicing of exon 5 (Figure 1).
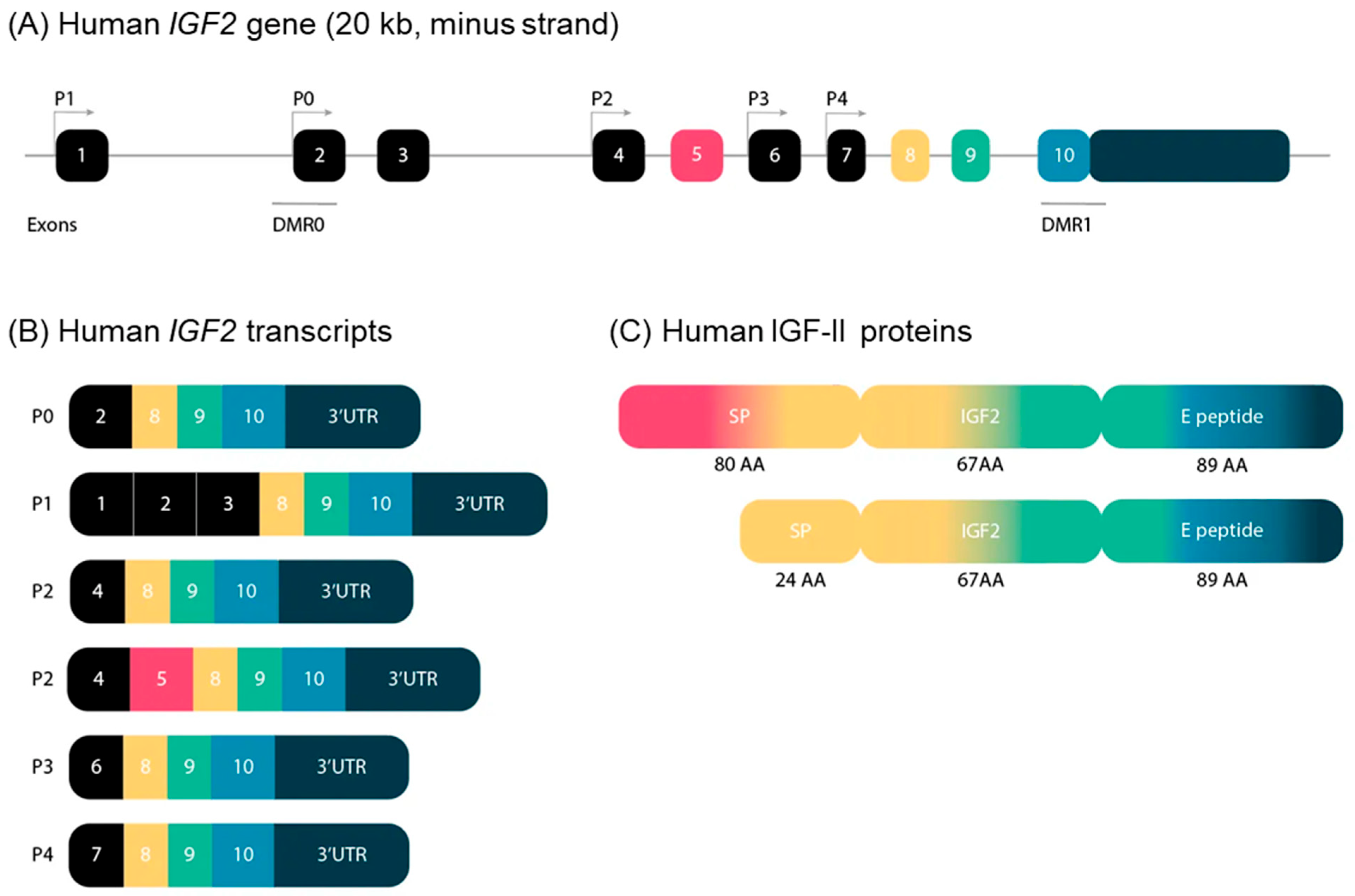
Figure 1. (A) Schematic representation of the structure of the IGF2 gene in humans. The IGF2 gene consists of 10 exons and is driven by five different promotors. The exons of the IGF2 gene are boxed. The black boxes indicate non-coding exons. The colored boxes indicate the coding exons. The turned arrows show the promotors (P) and indicate the transcription start sites. The blue lines indicate the differentially methylated regions (DMR) in the IGF2 gene. (B) Transcripts of the human IGF2 gene. IGF2 has six alternative transcripts, depending on promotors and splice sites used. (C) Human IGF-II proteins. IGF-II has two precursor proteins. Only exons 5, 8, 9 and 10 encode IGF-II proteins. Exon 5 is not included in the composition of the second precursor protein, which therefore has a smaller signal peptide. SP: signal peptide; AA: amino acids.
The human-secreted IGF-II is composed of 67 amino acids organized into four domains, the B, C, A, and D domains (listed in order from the N- to the C-terminus) [2]. Two types of protein precursors with different presumptive N-terminal signal peptides (consisting of 24 or 80 amino acids) give rise to mature human IGF-II, depending on the inclusion or exclusion of exon 5 in the IGF2 mRNA. The E peptide of the IGF-II precursor (also named “big” IGF-II), encoded by the 3-’end mRNA, is 89 amino acids long (Figure 1). It has been involved in paraneoplastic pancreatic independent hypoglycemia [3][4].
The 11p15.5 locus also includes H19, which is separated from IGF2 by about 80 kb (Figure 2). In both humans and mice, IGF2 and H19 are imprinted in a reciprocal manner. Moore et al. detected three Igf2 antisense transcripts relative to P0 transcription, with no open reading frames, in mice [5]. The exact role of these transcripts is unclear, and it is unknown whether their co-expression with the sense transcript (which has yet to be demonstrated), within the same cell, would influence mRNA stability.
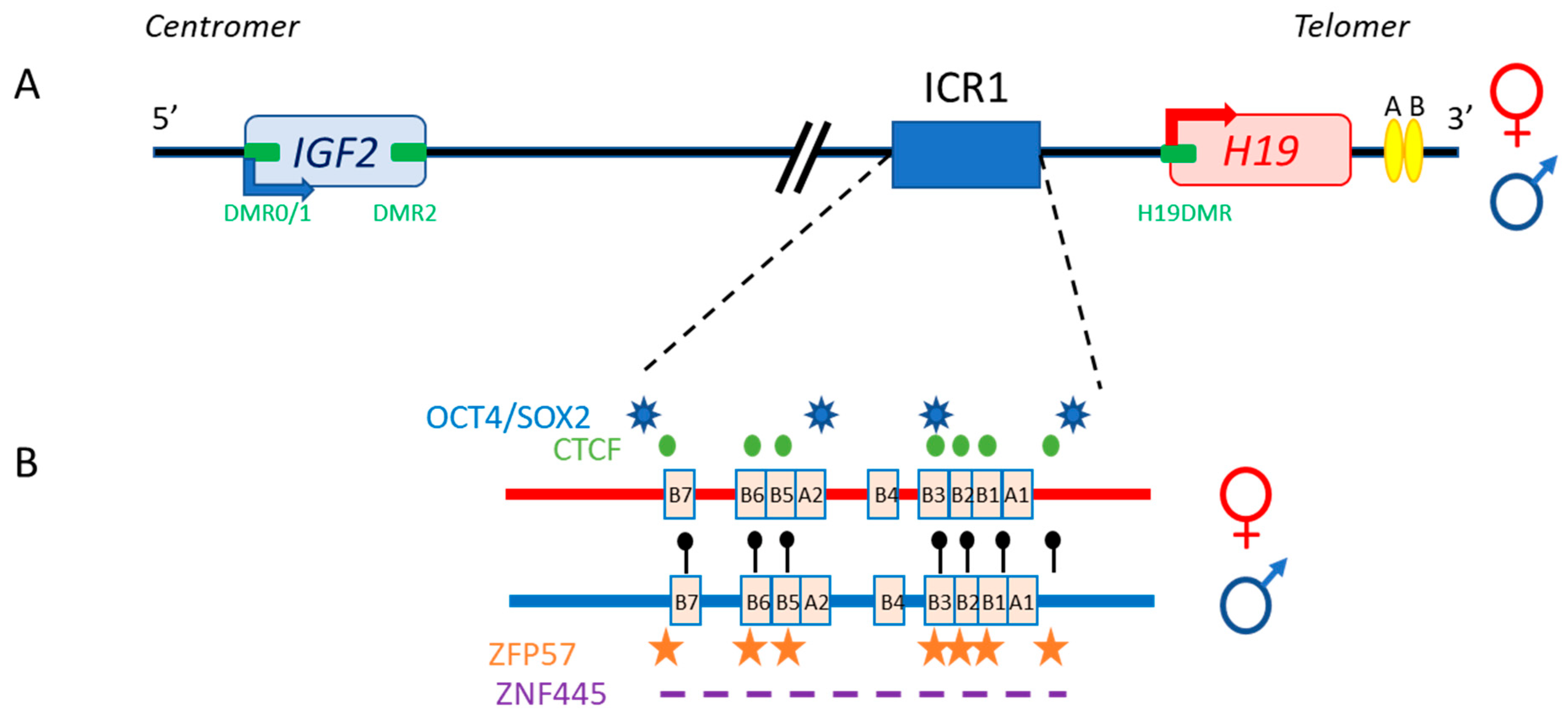
Figure 2. The human IGF2/H19 11p15.5 locus. (A) The IGF2 and H19 genes are separated by about 80 kb. IGF2 is paternally expressed (blue arrow), whereas H19 is maternally expressed (red arrow). The four DMR (green boxes) and enhancers (yellow ellipses) are represented. (B) H19/IGF2:IG-DMR (ICR1) in detail: OCT4/SOX2 (blue stars) and CTCF (green circles) binding sites on the maternal allele (red), and methylation sites (black lollipops), ZFP57 binding sites (orange stars) and the undefined ZNF445 binding site consensus sequence (purple dash) on the paternal allele (blue) are shown.
Different protein-binding sites are involved in regulating the expression of these two genes. The first evidence of differences in protein-binding sites and methylation status between the two alleles came from DNA hypersensitive site studies on the mouse Igf2/H19 locus [6]. These studies revealed clear differences in nuclease sensitivity between the parental chromosomes, with the presence of mutually exclusive hypersensitive sites (on the maternal chromosome) and DNA methylation sites (on the paternal chromosome).
The specific parent-of-origin pattern of expression of H19 and IGF2 at 11p15.5 is controlled by the allele-specific methylation status of H19/IGF2:IG-DMR. This locus is composed of seven CTCF-binding sites (CBS1-7) located in the A and B blocks of repeated domains (Figure 2). CTCF is a highly conserved zinc-finger DNA-binding protein with multiple roles in gene regulation [7]. The region orthologous to ICR1 in mouse contains only four CBS at the Igf2 locus. Several studies in humans have reported methylation at all CBS in H19/IGF2:IG-DMR on the paternal allele and established a correlation between the methylation of these CBS and IGF2 expression from the paternal allele [8]. Our team has also demonstrated homogeneous methylation levels for all CBS in humans [9]. In mice, CTCF binding to the unmethylated maternal ICR is essential for imprint maintenance in somatic cells, providing protection against aberrant de novo methylation at DMR throughout the locus. Furthermore, CTCF acts as an insulator, and CTCF binding creates a small loop of silent chromatin (CTCF binding to the maternal ICR regulates its interaction with matrix attachment region (MAR)3 and DMR1 at Igf2) (Figure 3), preventing enhancers from gaining access to the Igf2 promoter [10]. H19/IGF2:IG-DMR methylation on the paternal allele abolishes CTCF binding and ICR-mediated insulation, resulting in functional communication between promoters and enhancers, and an activation of Igf2 expression (see below).
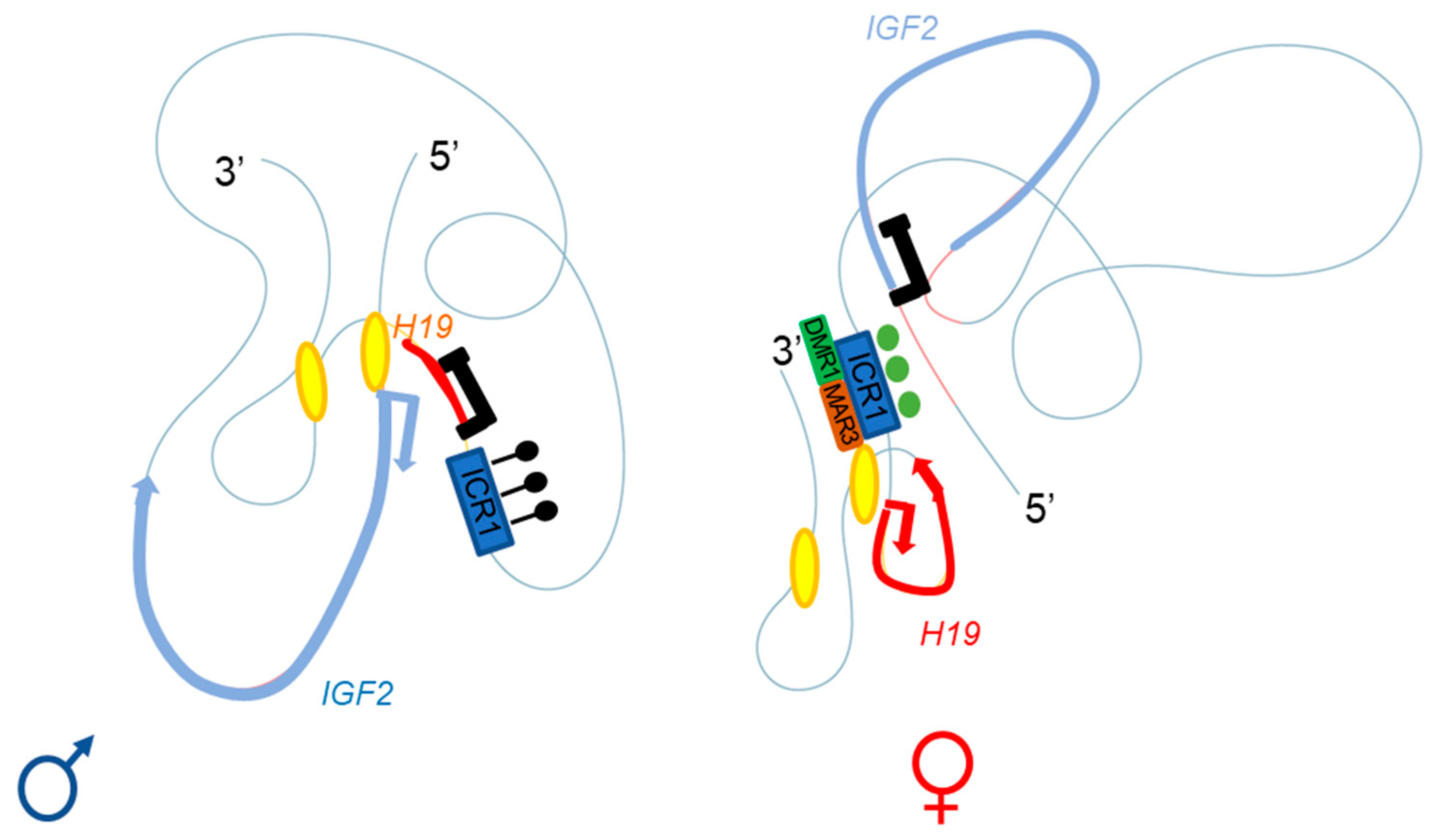
Figure 3. Adapted from Rovina et al. [Rovina, et al. Sci. Rep. 2020, 10, 8275]. Three-dimensional representation of the IGF2-H19 locus on the paternal (left) and maternal (right) chromosomes. The ICR1 of the paternal allele is methylated (black lollipops), with enhancers A and B (yellow ellipses) close to the IGF2 promoter; this conformation allows IGF2 expression and H19 repression. The CTCF of the maternal allele can bind the unmethylated ICR1 (green circles), regulating the interaction with DMR1 (green box) and the matrix attachment region (MAR)3 (brown box); the A and B (yellow ellipses) enhancers are close to the H19 promoter. This conformation allows H19 expression and IGF2 repression.
In addition to CBS, the human IGF2/H19 domain contains four binding sites for the pluripotency factors OCT4 and SOX2 [11]. There is evidence to suggest that the function of CTCF is modulated by the binding of OCT4/SOX2 to neighboring areas of DNA [12]. These factors play a role in maintaining or establishing the unmethylated status of the maternal allele. This hypothesis is strongly supported by in vitro experiments and by studies in transgenic mouse models showing that the OCT4/SOX2 binding sites in the maternal H19/IGF2:IG-DMR are essential for the full protection of DNA methylation during the establishment or maintenance phases [13][14]. Indeed, in humans, mutations of these OCT4/SOX2 binding sites in the maternal allele lead to hypermethylation of the CBS, followed by an increase in IGF2 expression leading to Beckwith–Wiedemann syndrome [15]. Two other factors, ZFP57 and ZNF445, protect ICR from DNA demethylation after fertilization. ZNF445 seems to be sufficient on its own in humans, whereas ZFP57 and ZNF445 cooperate in rodents (Figure 2) [16][17]. The IGF2/H19 domain also contains several other DMR: three within the IGF2 gene (DMR0, DMR1 and DMR2) and an additional DMR located in the H19 promoter (H19DMR), all of which are secondary DMR (somatic DMR that acquire their parent-specific DNA methylation mark in somatic diploid cells) methylated on the paternal allele [18] (Figure 2).
2. Three-Dimensional Organization
The regulation of IGF2 expression must be considered in three dimensions, with CTCF playing a major role in this aspect. There is experimental evidence to suggest that CTCF confers allele-specific effects on transcription via long-range chromatin interactions. The generation of these data was made possible by the emergence of 3C technology [19]. These interactions are dependent on the parental origin of the chromatin.
Series of deletions at the H19/Igf2 locus have made it possible to demonstrate the presence of several enhancers, two of which are predominantly endodermal and located 10 kb from the start site of the H19 transcript. These two enhancers target the H19 and Igf2 promoters, allowing expression of the corresponding genes (see below and Figure 2 and Figure 3) [20]. For the maternal allele, for which Igf2 expression is silent, 3C data are generally consistent with a model in which the CTCF-bound ICR contacts both the upstream DMR1 and a downstream matrix attachment region (MAR). Genetic studies have confirmed that CTCF binding to the ICR is required for both the formation of ICR-DMR1-MAR contacts and the prevention of maternal-specific enhancer-Igf2 promoter interactions. By contrast, on the paternal allele, which displays active Igf2 expression, all DMR sequences are methylated, preventing CTCF binding, and most of this region appears to be accessible, allowing more fluid contact with the enhancers [10][21].
Recent efforts to elucidate chromatin organization at the Igf2/H19 mouse locus, based on a combination of studies of allelic CTCF binding with both high-resolution and single-cell 3D chromatin organization assays, defined topologically associated domains (TAD) [22]. These studies determined the dynamic structure of the imprinted Igf2-H19 domain, and showed that CTCF binding occurred at multiple sites in both alleles, exclusively in ICR1 for the maternal. Furthermore, combinations of allelic 4C-seq and DNA-FISH revealed that CTCF binding to the paternal chromosome alone was correlated with a first level of sub-TAD structure. Additional CTCF binding to the differentially methylated region on the maternal chromosome adds a further layer of sub-TAD organization. This allele-specific sub-TAD organization may, thus, provide an instructive or permissive context for the correct activation of imprinted genes during development.
In humans, a specific parent-of-origin pattern of expression through TAD generation according to CTCF binding has been described, leading to IGF2 expression or silencing [23]. As in mice, the most important proteins for TAD architecture are CTCF (which bind to the ICR) (Figure 3). Moreover, crosstalk between IGF2/H19 and the CDKN1C/KCNQ1OT1 domain (another imprinted domain located in the same chromosome region) has been detected on the basis of a higher order of chromatin folding, suggesting the involvement of a mechanism for coordinating the expression of genes with the same expression status: IGF2 and KCNQ1OT1 on the paternal allele and H19 and CDKN1C on the maternal allele [23].
3. Trans-Regulation Mechanisms
In addition to being regulated by H19/IGF2:IG-DMR methylation and the three-dimensional organization of chromatin, IGF2 can be directly regulated through the activation of its promoters by several transcription factors, including those of the oncogenic HMGA2-PLAG1 pathway [24]. PLAG1 (pleiomorphic adenoma gene 1) overexpression was first observed in pleiomorphic adenomas of the salivary glands, identifying PLAG1 as an oncogene [25]. PLAG1 is a nuclear factor with seven zinc-finger domains that can bind IGF2 promoter P3, upregulating its transcriptional activity [25]. This finding has been confirmed in various other tumors, including hepatoblastomas, lipoblastomas and leukemia (review in [26]). Interestingly, Plag1 inactivation in mouse models results in pre- and postnatal growth retardation, despite an absence of change in Igf2 expression in embryos and pups [27].
HMGA2 (high mobility group AT-hook 2), initially named HMGI-C, is a member of the high-mobility group of proteins. Its expression is usually barely detectable in normal adult cells, but increases in cells transformed with viral oncogenes and in malignant tissues [28]. Mouse models of Hmga2 inactivation have a pygmy phenotype, with pre- and postnatal growth restriction and craniofacial abnormalities (a shortened head) [29]. The expression levels of HMGA2 and PLAG1 are highly correlated in thyroid tumors, and HMGA2 overexpression in cellular models is associated with an increase in PLAG1 expression [30].
The role of this oncogenic pathway in the control of IGF2 expression was highlighted in 2017, with the identification of additional mutations of HMGA2 and the first mutations of PLAG1 in patients referred for Silver–Russell syndrome (in addition to original mutations of IGF2). One of these PLAG1 mutations led to a downregulation of IGF2 expression in fibroblasts through a specific change in P3 promoter activity. Finally, the overexpression of HMGA2 and PLAG1 or their silencing in transfection assays result in a gain in expression or the downregulation of IGF2 expression, respectively [25].
DIS3L2 (DIS3-like 3′-5′ exoribonuclease 2) encodes a protein involved in the processing of mRNA and small non-coding RNAs. Homozygous loss-of-function variants of DIS3L2 lead to a rare condition called Perlman syndrome. This syndrome is characterized by excessive fetal growth and an increase in the risk of Wilms’ tumor [31]. In a mouse model, Dis3l2 invalidation was associated with an overexpression of Igf2 in nephron progenitor cells that was not associated with a loss of imprinting, as Igf2 still displayed monoallelic expression. The mechanism of Igf2 overexpression in this model remains to be determined [32].
Network of imprinted genes: In recent years, several studies in humans or animal models have shown that abnormalities at a given imprinted locus can impact at the expression of genes not only at the locus concerned, but also at other imprinted or non-imprinted loci [33][34][35][36]. This finding raised the possibility of an imprinted gene network, within which, imprinted genes are co-regulated. This pattern of regulation may partly account for the clinical overlap between imprinting disorders due to (epi)genetic defects at different imprinted loci [37]. For example, a strong clinical overlap between Silver–Russell syndrome (SRS, OMIM #180860) and Temple syndrome (TS14, OMIM #616222) has been described, despite the existence of several syndrome-specific traits, including pre- and postnatal growth restriction, relative macrocephaly, feeding difficulties and a protruding forehead [38]. IGF2 downregulation is thought to be the molecular mechanism underlying the SRS phenotype, with about 40% of SRS patients presenting hypomethylation at the H19/IGF2:IG-DMR [39]. TS14 is mostly due to abnormalities of the imprinted 14q32.2 locus. This locus contains non-coding RNA sequences that are expressed from the maternal allele only (including the two long non-coding RNA, MEG3 and MEG8). In cases of maternal uniparental disomy of chromosome 14 or hypomethylation at the MEG3/DLK1:IG-DMR, MEG3 and MEG8 are expressed from both the paternal and maternal alleles, leading to an increase in the level of expression of these two genes. Abnormally low levels of IGF2 expression have been reported in the fibroblasts of TS14 patients, despite normal H19/IGF2:IG-DMR methylation. Furthermore, in control fibroblasts, the overexpression of MEG3 and MEG8 leads to a downregulation of IGF2. Conversely, the silencing of MEG3 and/or MEG8 in control fibroblasts leads to an upregulation of IGF2 expression. Thus, MEG3 and MEG8, which are expressed from the maternal 14q32.2 locus, regulates IGF2 expression at 11p15.5, providing support for the hypothesis of an imprinted gene network [40].
This entry is adapted from the peer-reviewed paper 10.3390/cells11121886
References
- Baral, K.; Rotwein, P. The Insulin-like Growth Factor 2 Gene in Mammals: Organizational Complexity within a Conserved Locus. PLoS ONE 2019, 14, e0219155.
- De Pagter-Holthuizen, P.; Jansen, M.; van Schaik, F.M.A.; van der Kammen, R.; Oosterwijk, C.; Van den Brande, J.L.; Sussenbach, J.S. The Human Insulin-like Growth Factor II Gene Contains Two Development-Specific Promoters. FEBS Lett. 1987, 214, 259–264.
- Zapf, J.; Futo, E.; Peter, M.; Froesch, E.R. Can “Big” Insulin-like Growth Factor II in Serum of Tumor Patients Account for the Development of Extrapancreatic Tumor Hypoglycemia? J. Clin. Investig. 1992, 90, 2574–2584.
- Rotwein, P. Large-Scale Analysis of Variation in the Insulin-like Growth Factor Family in Humans Reveals Rare Disease Links and Common Polymorphisms. J. Biol. Chem. 2017, 292, 9252–9261.
- Moore, T.; Constancia, M.; Zubair, M.; Bailleul, B.; Feil, R.; Sasaki, H.; Reik, W. Multiple Imprinted Sense and Antisense Transcripts, Differential Methylation and Tandem Repeats in a Putative Imprinting Control Region Upstream of Mouse Igf2. Proc. Natl. Acad. Sci. USA 1997, 94, 12509–12514.
- Khosla, S.; Aitchison, A.; Gregory, R.; Allen, N.D.; Feil, R. Parental Allele-Specific Chromatin Configuration in a Boundary-Imprinting-Control Element Upstream of the Mouse H19 Gene. Mol. Cell. Biol. 1999, 19, 2556–2566.
- Phillips, J.E.; Corces, V.G. CTCF: Master Weaver of the Genome. Cell 2009, 137, 1194–1211.
- Takai, D.; Gonzales, F.A.; Tsai, Y.C.; Thayer, M.J.; Jones, P.A. Large Scale Mapping of Methylcytosines in CTCF-Binding Sites in the Human H19 Promoter and Aberrant Hypomethylation in Human Bladder Cancer. Hum. Mol. Genet. 2001, 10, 2619–2626.
- Abi Habib, W.; Azzi, S.; Brioude, F.; Steunou, V.; Thibaud, N.; Das Neves, C.; Le Jule, M.; Chantot-Bastaraud, S.; Keren, B.; Lyonnet, S.; et al. Extensive Investigation of the IGF2/H19 Imprinting Control Region Reveals Novel OCT4/SOX2 Binding Site Defects Associated with Specific Methylation Patterns in Beckwith-Wiedemann Syndrome. Hum. Mol. Genet. 2014, 23, 5763–5773.
- Kurukuti, S.; Tiwari, V.K.; Tavoosidana, G.; Pugacheva, E.; Murrell, A.; Zhao, Z.; Lobanenkov, V.; Reik, W.; Ohlsson, R. CTCF Binding at the H19 Imprinting Control Region Mediates Maternally Inherited Higher-Order Chromatin Conformation to Restrict Enhancer Access to Igf2. Proc. Natl. Acad. Sci. USA 2006, 103, 10684–10689.
- Demars, J.; Shmela, M.E.; Rossignol, S.; Okabe, J.; Netchine, I.; Azzi, S.; Cabrol, S.; Le Caignec, C.; David, A.; Le Bouc, Y.; et al. Analysis of the IGF2/H19 Imprinting Control Region Uncovers New Genetic Defects, Including Mutations of OCT-Binding Sequences, in Patients with 11p15 Fetal Growth Disorders. Hum. Mol. Genet. 2010, 19, 803–814.
- Weth, O.; Renkawitz, R. CTCF Function Is Modulated by Neighboring DNA Binding Factors. Biochem. Cell Biol. 2011, 89, 459–468.
- Zimmerman, D.L.; Boddy, C.S.; Schoenherr, C.S. Oct4/Sox2 Binding Sites Contribute to Maintaining Hypomethylation of the Maternal Igf2/H19 Imprinting Control Region. PLoS ONE 2013, 8, e81962.
- Sakaguchi, R.; Okamura, E.; Matsuzaki, H.; Fukamizu, A.; Tanimoto, K. Sox-Oct Motifs Contribute to Maintenance of the Unmethylated H19 ICR in YAC Transgenic Mice. Hum. Mol. Genet. 2013, 22, 4627–4637.
- Abi Habib, W.; Brioude, F.; Azzi, S.; Salem, J.; Das Neves, C.; Personnier, C.; Chantot-Bastaraud, S.; Keren, B.; Le Bouc, Y.; Harbison, M.D.; et al. 11p15 ICR1 Partial Deletions Associated with IGF2/H19 DMR Hypomethylation and Silver-Russell Syndrome. Hum. Mutat. 2017, 38, 105–111.
- Quenneville, S.; Verde, G.; Corsinotti, A.; Kapopoulou, A.; Jakobsson, J.; Offner, S.; Baglivo, I.; Pedone, P.V.; Grimaldi, G.; Riccio, A.; et al. In Embryonic Stem Cells, ZFP57/KAP1 Recognize a Methylated Hexanucleotide to Affect Chromatin and DNA Methylation of Imprinting Control Regions. Mol. Cell 2011, 44, 361–372.
- Takahashi, N.; Coluccio, A.; Thorball, C.W.; Planet, E.; Shi, H.; Offner, S.; Turelli, P.; Imbeault, M.; Ferguson-Smith, A.C.; Trono, D. ZNF445 Is a Primary Regulator of Genomic Imprinting. Genes Dev. 2019, 33, 49–54.
- Monk, D.; Sanches, R.; Arnaud, P.; Apostolidou, S.; Hills, F.A.; Abu-Amero, S.; Murrell, A.; Friess, H.; Reik, W.; Stanier, P.; et al. Imprinting of IGF2 P0 Transcript and Novel Alternatively Spliced INS-IGF2 Isoforms Show Differences between Mouse and Human. Hum. Mol. Genet. 2006, 15, 1259–1269.
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing Chromosome Conformation. Science 2002, 295, 1306–1311.
- Arney, K.L. H19 and Igf2—Enhancing the Confusion? Trends Genet. 2003, 19, 17–23.
- Yoon, Y.S.; Jeong, S.; Rong, Q.; Park, K.-Y.; Chung, J.H.; Pfeifer, K. Analysis of the H19ICR Insulator. Mol. Cell. Biol. 2007, 27, 3499–3510.
- Llères, D.; Moindrot, B.; Pathak, R.; Piras, V.; Matelot, M.; Pignard, B.; Marchand, A.; Poncelet, M.; Perrin, A.; Tellier, V.; et al. CTCF Modulates Allele-Specific Sub-TAD Organization and Imprinted Gene Activity at the Mouse Dlk1-Dio3 and Igf2-H19 Domains. Genome Biol. 2019, 20, 272.
- Rovina, D.; La Vecchia, M.; Cortesi, A.; Fontana, L.; Pesant, M.; Maitz, S.; Tabano, S.; Bodega, B.; Miozzo, M.; Sirchia, S.M. Profound Alterations of the Chromatin Architecture at Chromosome 11p15.5 in Cells from Beckwith-Wiedemann and Silver-Russell Syndromes Patients. Sci. Rep. 2020, 10, 8275.
- Abi Habib, W.; Brioude, F.; Edouard, T.; Bennett, J.T.; Lienhardt-Roussie, A.; Tixier, F.; Salem, J.; Yuen, T.; Azzi, S.; Le Bouc, Y.; et al. Genetic Disruption of the Oncogenic HMGA2-PLAG1-IGF2 Pathway Causes Fetal Growth Restriction. Genet. Med. 2018, 20, 250–258.
- Voz, M.L.; Agten, N.S.; Van de Ven, W.J.; Kas, K. PLAG1, the Main Translocation Target in Pleomorphic Adenoma of the Salivary Glands, Is a Positive Regulator of IGF-II. Cancer Res. 2000, 60, 106–113.
- Van Dyck, F.; Declercq, J.; Braem, C.V.; Van de Ven, W.J.M. PLAG1, the Prototype of the PLAG Gene Family: Versatility in Tumour Development. Int. J. Oncol. 2007, 30, 765–774.
- Hensen, K.; Braem, C.; Declercq, J.; Van Dyck, F.; Dewerchin, M.; Fiette, L.; Denef, C.; Van de Ven, W.J.M. Targeted Disruption of the Murine Plag1 Proto-Oncogene Causes Growth Retardation and Reduced Fertility. Dev. Growth Differ. 2004, 46, 459–470.
- Manfioletti, G.; Giancotti, V.; Bandiera, A.; Buratti, E.; Sautière, P.; Cary, P.; Crane-Robinson, C.; Coles, B.; Goodwin, G.H. CDNA Cloning of the HMGI-C Phosphoprotein, a Nuclear Protein Associated with Neoplastic and Undifferentiated Phenotypes. Nucleic Acids Res. 1991, 19, 6793–6797. [Google Scholar] [CrossRef]
- Zhou, X.; Benson, K.F.; Ashar, H.R.; Chada, K. Mutation Responsible for the Mouse Pygmy Phenotype in the Developmentally Regulated Factor HMGI-C. Nature 1995, 376, 771–774. [Google Scholar] [CrossRef]
- Klemke, M.; Müller, M.H.; Wosniok, W.; Markowski, D.N.; Nimzyk, R.; Helmke, B.M.; Bullerdiek, J. Correlated Expression of HMGA2 and PLAG1 in Thyroid Tumors, Uterine Leiomyomas and Experimental Models. PLoS ONE 2014, 9, e88126. [Google Scholar] [CrossRef]
- Astuti, D.; Morris, M.R.; Cooper, W.N.; Staals, R.H.J.; Wake, N.C.; Fews, G.A.; Gill, H.; Gentle, D.; Shuib, S.; Ricketts, C.J.; et al. Germline Mutations in DIS3L2 Cause the Perlman Syndrome of Overgrowth and Wilms Tumor Susceptibility. Nat. Genet. 2012, 44, 277–284. [Google Scholar] [CrossRef]
- Hunter, R.W.; Liu, Y.; Manjunath, H.; Acharya, A.; Jones, B.T.; Zhang, H.; Chen, B.; Ramalingam, H.; Hammer, R.E.; Xie, Y.; et al. Loss of Dis3l2 Partially Phenocopies Perlman Syndrome in Mice and Results in Up-Regulation of Igf2 in Nephron Progenitor Cells. Genes Dev. 2018, 32, 903–908. [Google Scholar] [CrossRef]
- Varrault, A.; Gueydan, C.; Delalbre, A.; Bellmann, A.; Houssami, S.; Aknin, C.; Severac, D.; Chotard, L.; Kahli, M.; Le Digarcher, A.; et al. Zac1 Regulates an Imprinted Gene Network Critically Involved in the Control of Embryonic Growth. Dev. Cell 2006, 11, 711–722. [Google Scholar] [CrossRef]
- Gabory, A.; Ripoche, M.-A.; Le Digarcher, A.; Watrin, F.; Ziyyat, A.; Forné, T.; Jammes, H.; Ainscough, J.F.X.; Surani, M.A.; Journot, L.; et al. H19 Acts as a Trans Regulator of the Imprinted Gene Network Controlling Growth in Mice. Development 2009, 136, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Al Adhami, H.; Evano, B.; Le Digarcher, A.; Gueydan, C.; Dubois, E.; Parrinello, H.; Dantec, C.; Bouschet, T.; Varrault, A.; Journot, L. A Systems-Level Approach to Parental Genomic Imprinting: The Imprinted Gene Network Includes Extracellular Matrix Genes and Regulates Cell Cycle Exit and Differentiation. Genome Res. 2015, 25, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Whipple, A.J.; Breton-Provencher, V.; Jacobs, H.N.; Chitta, U.K.; Sur, M.; Sharp, P.A. Imprinted Maternally Expressed MicroRNAs Antagonize Paternally Driven Gene Programs in Neurons. Mol. Cell 2020, 78, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Eggermann, T.; Davies, J.H.; Tauber, M.; van den Akker, E.; Hokken-Koelega, A.; Johansson, G.; Netchine, I. Growth Restriction and Genomic Imprinting-Overlapping Phenotypes Support the Concept of an Imprinting Network. Genes 2021, 12, 585. [Google Scholar] [CrossRef] [PubMed]
- Geoffron, S.; Abi Habib, W.; Chantot-Bastaraud, S.; Dubern, B.; Steunou, V.; Azzi, S.; Afenjar, A.; Busa, T.; Pinheiro Canton, A.; Chalouhi, C.; et al. Chromosome 14q32.2 Imprinted Region Disruption as an Alternative Molecular Diagnosis of Silver-Russell Syndrome. J. Clin. Endocrinol. Metab. 2018, 103, 2436–2446. [Google Scholar] [CrossRef]
- Wakeling, E.L.; Brioude, F.; Lokulo-Sodipe, O.; O’Connell, S.M.; Salem, J.; Bliek, J.; Canton, A.P.M.; Chrzanowska, K.H.; Davies, J.H.; Dias, R.P.; et al. Diagnosis and Management of Silver-Russell Syndrome: First International Consensus Statement. Nat. Rev. Endocrinol. 2017, 13, 105–124. [Google Scholar] [CrossRef]
- Abi Habib, W.; Brioude, F.; Azzi, S.; Rossignol, S.; Linglart, A.; Sobrier, M.-L.; Giabicani, É.; Steunou, V.; Harbison, M.D.; Le Bouc, Y.; et al. Transcriptional Profiling at the DLK1/MEG3 Domain Explains Clinical Overlap between Imprinting Disorders. Sci. Adv. 2019, 5, eaau9425.
This entry is offline, you can click here to edit this entry!
