Polyploidy events have long been recognized as the primary driving force behind the survival of the vast majority of plant lineages, playing critical roles in crop evolution, speciation, and domestication. The fascinating modern genomics era has revealed that the genomes of all flowering plants have been polyploidized multiple times. To help safeguard the future of the global food supply in the face of climate change, a thorough understanding of polyploid genomes is critical, especially for the improvement of members of the grass family, Poaceae, which includes the world’s big three cereals—rice, wheat, and maize—as well as some potential underutilized ancient grasses (such as teff) that are less well known or studied.
- climate change
- evolution
- food security
- plant breeding
- plant diversity
- polyploids
1. Introduction
2. Impacts of Polyploidy at a Glance
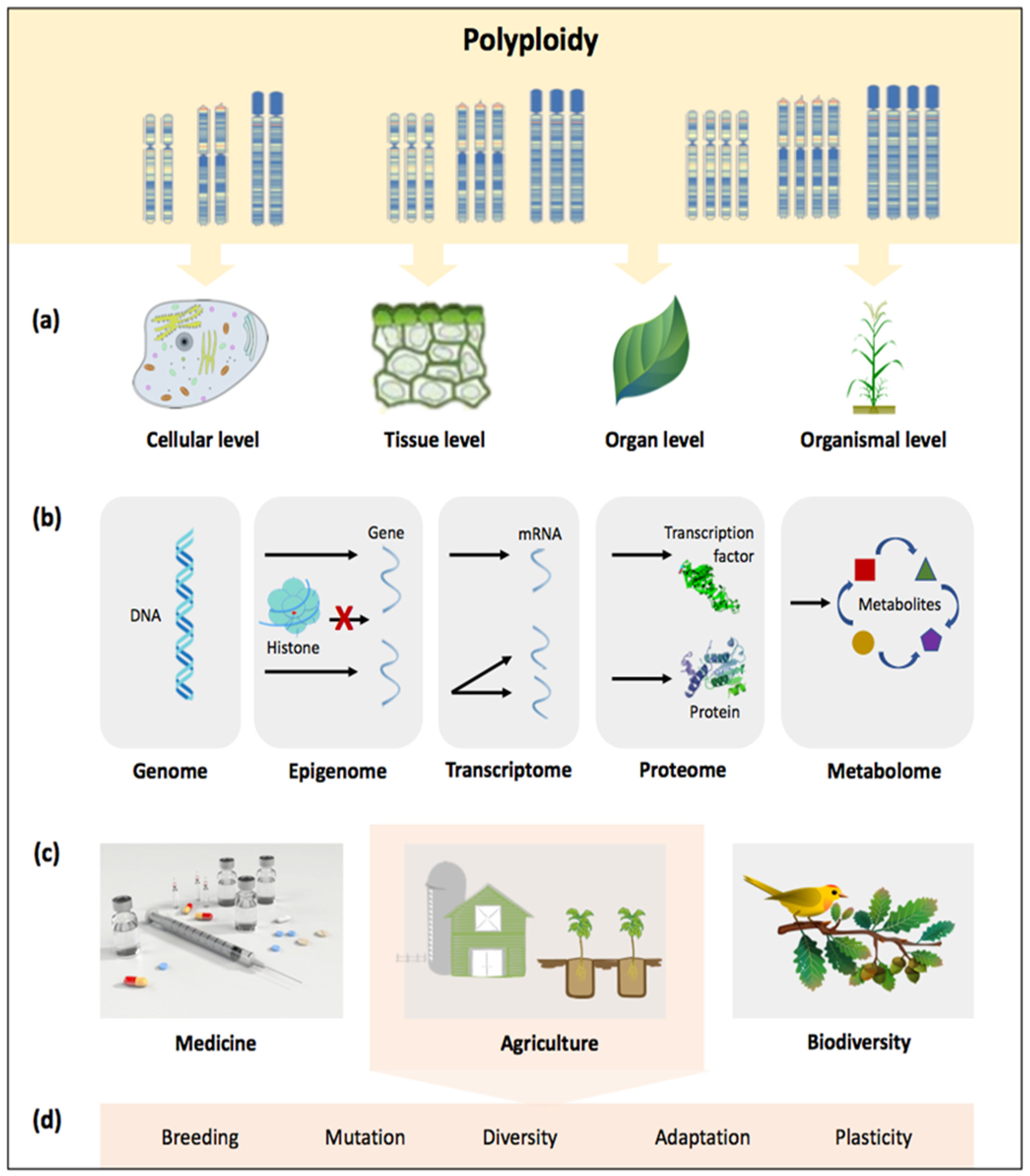
2.1. Diversity and Evolutionary Dynamics
2.2. Crop Improvement
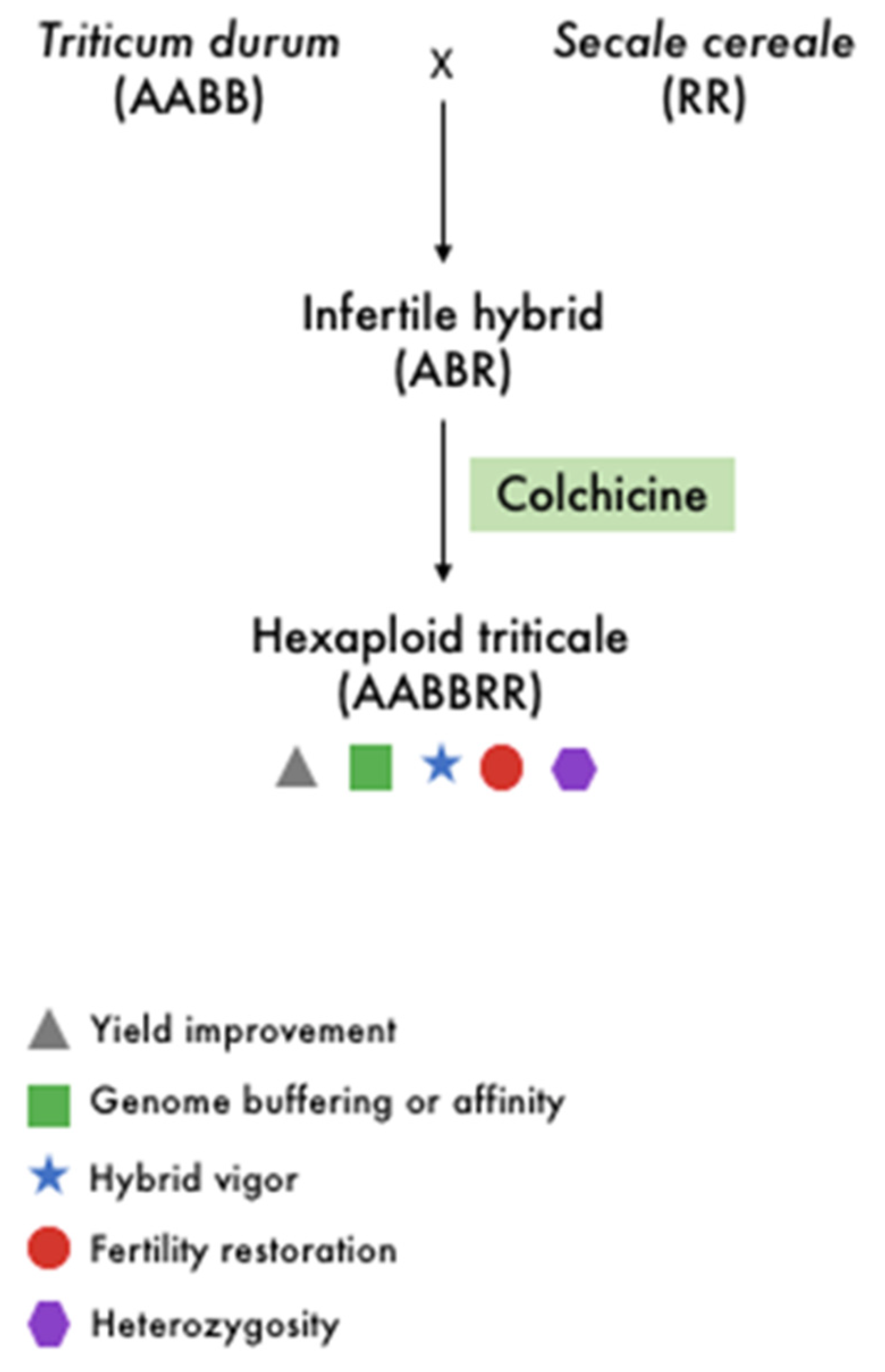
3. Dissecting the Polyploid Potential of Current and Future Crops in the Genomic Era
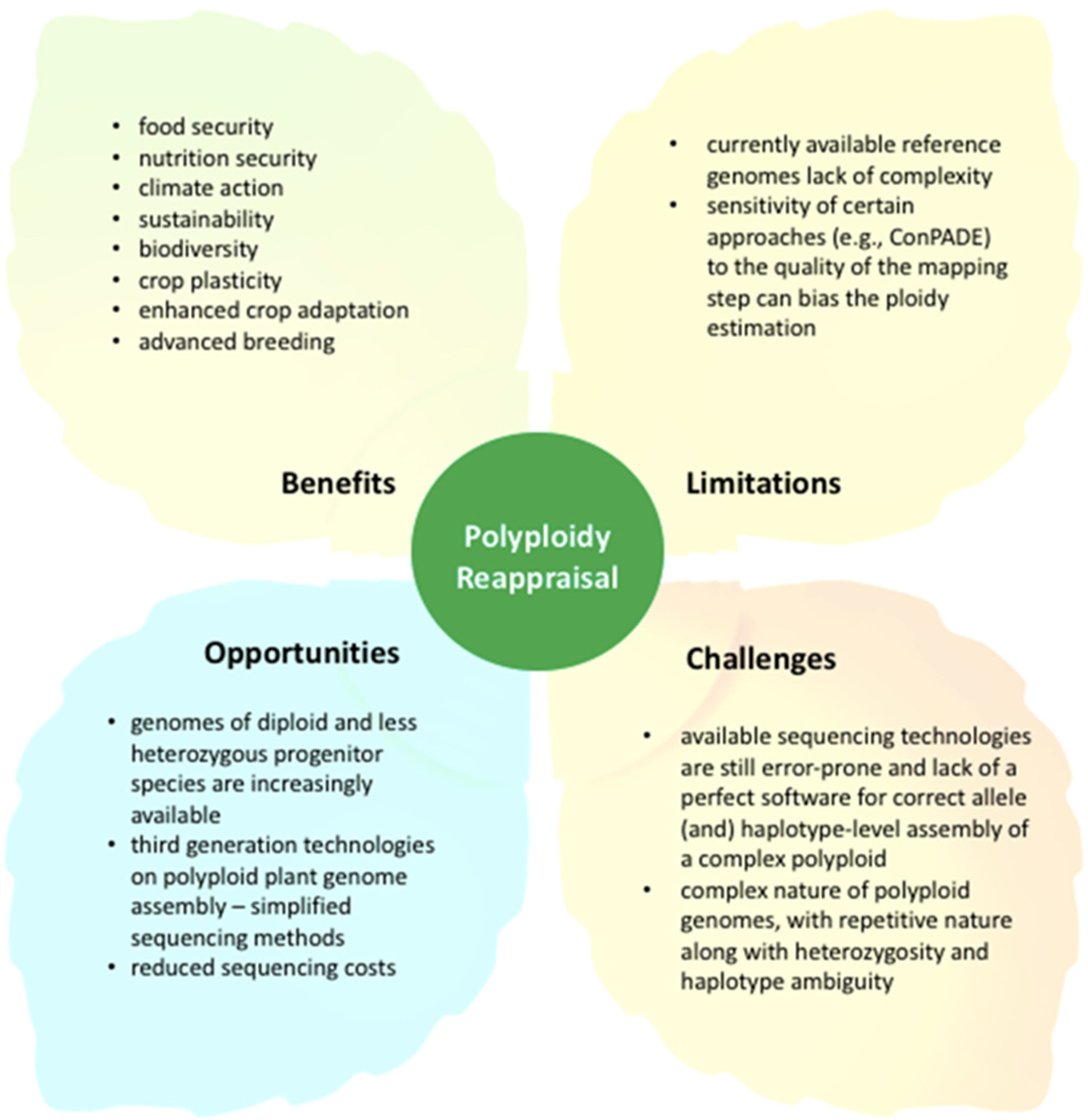
4. Reappraising Polyploidy for Crop Improvement in the 21st Century: The Road Ahead
This entry is adapted from the peer-reviewed paper 10.3390/biology11050636
References
- National Research Council (US). 4, Cretaceous-Tertiary (K/T) Mass Extinction: Effect of Global Change on Calcareous Microplankton. In Panel on Effects of Past Global Change on Life. Effects of Past Global Change on Life; National Academies Press (US): Washington, DC, USA, 1995; pp. 72–93.
- Chiarenza, A.A.; Farnsworth, A.; Mannion, P.D.; Lunt, D.J.; Valdes, P.J.; Morgan, J.V.; Allison, P.A. Asteroid impact, not volcanism, caused the end-Cretaceous dinosaur extinction. Proc. Natl. Acad. Sci. USA 2020, 117, 17084–17093.
- Carvalho, M.R.; Jaramillo, C.; de la Parra, F.; Caballero-Rodríguez, D.; Herrera, F.; Wing, S.; Turner, B.L.; D’Apolito, C.; Romero-Baez, M.; Narvaez, P. Extinction at the end-Cretaceous and the origin of modern tropical rainforests. Science 2021, 372, 63–68.
- Fawcett, J.A.; Maere, S.; Van De Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742.
- Fox, D.T.; Soltis, D.E.; Soltis, P.S.; Ashman, T.-L.; Van de Peer, Y. Polyploidy: A biological force from cells to ecosystems. Trends Cell. Biol. 2020, 30, 688–694.
- Wendel, J.; Doyle, J. Plant diversity and evolution: Genotypic and phenotypic variation in higher plants. In Polyploidy and Evolution in Plants; Henry, R.J., Ed.; CABI Publishing: Wallington, UK, 2005; pp. 97–117.
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71.
- Kyriakidou, M.; Tai, H.H.; Anglin, N.L.; Ellis, D.; Strömvik, M.V. Current strategies of polyploid plant genome sequence assembly. Front. Plant. Sci. 2018, 9, 1660.
- Van de Peer, Y.; Ashman, T.-L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26.
- Renny-Byfield, S.; Wendel, J.F. Doubling down on genomes: Polyploidy and crop plants. Am. J. Bot. 2014, 101, 1711–1725.
- Walden, N.; German, D.A.; Wolf, E.M.; Kiefer, M.; Rigault, P.; Huang, X.-C.; Kiefer, C.; Schmickl, R.; Franzke, A.; Neuffer, B. Nested whole-genome duplications coincide with diversification and high morphological disparity in Brassicaceae. Nat. Commun. 2020, 11, 3795.
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801.
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141.
- Scholes, D.R.; Paige, K.N. Plasticity in ploidy: A generalized response to stress. Trends Plant Sci. 2015, 20, 165–175.
- Levin, D.A. Polyploidy and novelty in flowering plants. Am. Nat. 1983, 122, 1–25.
- Doyle, J.J.; Coate, J.E. Polyploidy, the nucleotype, and novelty: The impact of genome doubling on the biology of the cell. Int. J. Plant Sci. 2019, 180, 1–52.
- Koenen, E.J.; Ojeda, D.I.; Bakker, F.T.; Wieringa, J.J.; Kidner, C.; Hardy, O.J.; Pennington, R.T.; Herendeen, P.S.; Bruneau, A.; Hughes, C.E. The origin of the legumes is a complex paleopolyploid phylogenomic tangle closely associated with the Cretaceous–Paleogene (K–Pg) mass extinction event. Syst. Biol. 2021, 70, 508–526.
- Conover, J.L.; Karimi, N.; Stenz, N.; Ané, C.; Grover, C.E.; Skema, C.; Tate, J.A.; Wolff, K.; Logan, S.A.; Wendel, J.F. A Malvaceae mystery: A mallow maelstrom of genome multiplications and maybe misleading methods? J. Integr. Plant Biol. 2019, 61, 12–31.
- Meyer, R.S.; Choi, J.Y.; Sanches, M.; Plessis, A.; Flowers, J.M.; Amas, J.; Dorph, K.; Barretto, A.; Gross, B.; Fuller, D.Q. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat. Genet. 2016, 48, 1083–1088.
- Woodhouse, M.; Hufford, M. Parallelism and convergence in post-domestication adaptation in cereal grasses. Philos. Trans. R. Soc. B. 2019, 374, 20180245.
- Bennetzin, J.L.; Freeling, M. Grasses as a single genetic system: Genome composition, collinearity and compatibility. Trends Genet. 1993, 9, 259–261.
- Paterson, A.; Bowers, J.; Chapman, B. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 2004, 101, 9903–9908.
- Ozkan, H.; Levy, A.A.; Feldman, M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell 2001, 13, 1735–1747.
- Chantret, N.; Salse, J.; Sabot, F.; Rahman, S.; Bellec, A.; Laubin, B.; Dubois, I.; Dossat, C.; Sourdille, P.; Joudrier, P. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 2005, 17, 1033–1045.
- Zhang, Z.; Belcram, H.; Gornicki, P.; Charles, M.; Just, J.; Huneau, C.; Magdelenat, G.; Couloux, A.; Samain, S.; Gill, B.S. Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 18737–18742.
- Yang, C.; Yang, Z.; Zhao, L.; Sun, F.; Liu, B. A newly formed hexaploid wheat exhibits immediate higher tolerance to nitrogen-deficiency than its parental lines. BMC Plant Biol. 2018, 18, 113.
- Spor, A.; Roucou, A.; Mounier, A.; Bru, D.; Breuil, M.-C.; Fort, F.; Vile, D.; Roumet, P.; Philippot, L.; Violle, C. Domestication-driven changes in plant traits associated with changes in the assembly of the rhizosphere microbiota in tetraploid wheat. Science 2020, 10, 12234.
- Di Vittori, V.; Gioia, T.; Rodriguez, M.; Bellucci, E.; Bitocchi, E.; Nanni, L.; Attene, G.; Rau, D.; Papa, R. Convergent evolution of the seed shattering trait. Genes 2019, 10, 68.
- Giraud, D.; Lima, O.; Rousseau-Gueutin, M.; Salmon, A.; Ainouche, M. Gene and transposable element expression evolution following recent and past polyploidy events in Spartina (Poaceae). Front. Genet. 2021, 12, 589160.
- Rutland, C.A.; Hall, N.D.; McElroy, J.S. The impact of polyploidization on the evolution of weed species: Historical understanding and current limitations. Front. Agron. 2021, 3, 5.
- Nieto Feliner, G.; Casacuberta, J.; Wendel, J.F. Genomics of evolutionary novelty in hybrids and polyploids. Front. Genet. 2020, 11, 792.
- Fort, A.; Ryder, P.; McKeown, P.C.; Wijnen, C.; Aarts, M.G.; Sulpice, R.; Spillane, C. Disaggregating polyploidy, parental genome dosage and hybridity contributions to heterosis in Arabidopsis thaliana. New Phytol. 2016, 209, 590–599.
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS meets germplasm. Plant Biotechnol. J. 2020, 18, 1507–1525.
- Labroo, M.R.; Studer, A.J.; Rutkoski, J.E. Heterosis and hybrid crop breeding: A multidisciplinary review. Front. Genet. 2021, 12, 234.
- Karbstein, K.; Rahmsdorf, E.; Tomasello, S.; Hodač, L.; Hörandl, E. Moving beyond assumptions: Polyploidy and environmental effects explain a geographical parthenogenesis scenario in European plants. Mol. Ecol. 2021, 30, 2659–2675.
- Silkova, O.G.; Ivanova, Y.N.; Loginova, D.B.; Solovey, L.A.; Sycheva, E.A.; Dubovets, N.I. Karyotype Reorganization in Wheat–Rye Hybrids Obtained via Unreduced Gametes: Is There a Limit to the Chromosome Number in Triticale? Plants 2021, 10, 2052.
- Koide, Y.; Kuniyoshi, D.; Kishima, Y. Fertile tetraploids: New resources for future rice breeding? Front. Plant Sci. 2020, 11, 1231.
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125.
- Jiao, W.-B.; Schneeberger, K. The impact of third generation genomic technologies on plant genome assembly. Curr. Opin. Plant Biol. 2017, 36, 64–70.
- Garsmeur, O.; Droc, G.; Antonise, R.; Grimwood, J.; Potier, B.; Aitken, K.; Jenkins, J.; Martin, G.; Charron, C.; Hervouet, C. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 2018, 9, 2638.
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312.
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573.
- Zhang, K.; Wang, X.; Cheng, F. Plant polyploidy: Origin, evolution, and its influence on crop domestication. Hortic. Plant J. 2019, 5, 231–239.
- Salman-Minkov, A.; Sabath, N.; Mayrose, I. Whole-genome duplication as a key factor in crop domestication. Nat. Plants 2016, 2, 16115.
- Bharadwaj, D.N. Polyploidy in crop improvement and evolution. In Plant Biology Biotechnology; Springer: New Delhi, India, 2015; pp. 619–638.
- Schoenfelder, K.P.; Fox, D.T. The expanding implications of polyploidy. J. Cell. Biol. 2015, 209, 485.
- Paige, K.N. Overcompensation, environmental stress, and the role of endoreduplication. Am. J. Bot. 2018, 105, 1105–1108.
- Sethuraman, G.; Mohd Zain, N.A.; Yusoff, S.; Ng, Y.M.; Baisakh, N.; Cheng, A. Revamping Ecosystem Services through Agroecology—The Case of Cereals. Agriculture 2021, 11, 204.
